9CTP
 
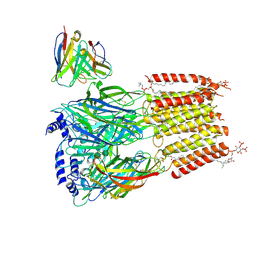 | | Native human GABAA receptor of beta2-alpha1-beta2-alpha3-gamma2 assembly | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2024-07-25 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Resolving native GABA A receptor structures from the human brain.
Nature, 638, 2025
|
|
5U23
 
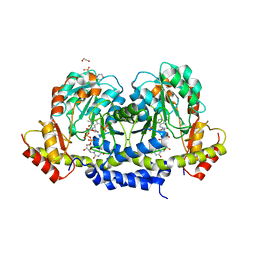 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
7HDH
 
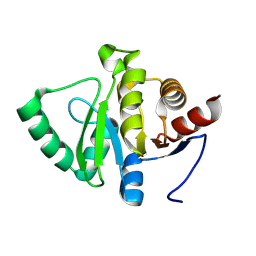 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003726 | | Descriptor: | Non-structural protein 3, [(2R)-5,5-dimethyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-2-yl]methanol, [(2S)-5,5-dimethyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-2-yl]methanol | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
4KDL
 
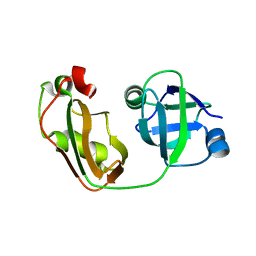 | | Crystal structure of p97/VCP N in complex with OTU1 UBXL | | Descriptor: | Transitional endoplasmic reticulum ATPase, Ubiquitin thioesterase OTU1 | | Authors: | Kim, S.J, Kim, E.E. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Ovarian Tumor Domain-containing Protein 1 (OTU1) Binding to p97/Valosin-containing Protein (VCP).
J.Biol.Chem., 289, 2014
|
|
4FWL
 
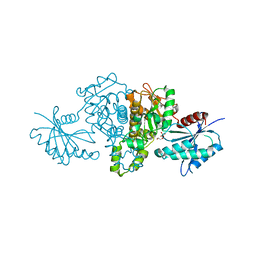 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with Phosphate (PO4) | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
5FCK
 
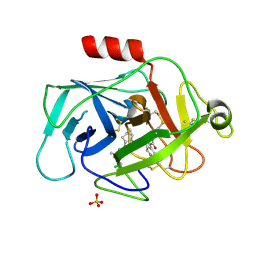 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[[(1~{R})-1-(3-chloranyl-2-fluoranyl-phenyl)ethyl]carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]pyrazolo[3,4-c]pyridine-3-carboxamide, Complement factor D, SULFATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
4NMS
 
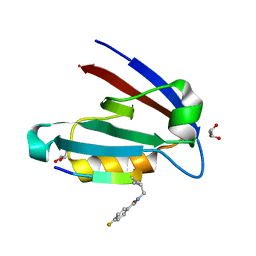 | |
3IID
 
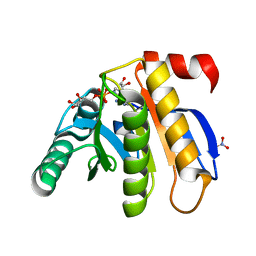 | | Crystal structure of the macro domain of human histone macroH2A1.1 in complex with ADP-ribose (form A) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Core histone macro-H2A.1, Isoform 1, ... | | Authors: | Hothorn, M, Bortfeld, M, Ladurner, A.G, Scheffzek, K. | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6Q1P
 
 | | Glucocerebrosidase in complex with pharmacological chaperone norIMX8 | | Descriptor: | (2S,3S,4S,5R)-2-{[(4-methylpentyl)sulfonyl]methyl}piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
7HEX
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003733 | | Descriptor: | DIMETHYL SULFOXIDE, Non-structural protein 3, [(2S,5R)-5-(1,3-dimethyl-1H-pyrazol-4-yl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-2-yl]methanol | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
3AGK
 
 | | Crystal structure of archaeal translation termination factor, aRF1 | | Descriptor: | Peptide chain release factor subunit 1 | | Authors: | Kobayashi, K, Kikuno, I, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Omnipotent role of archaeal elongation factor 1 alpha (EF1{alpha}) in translational elongation and termination, and quality control of protein synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7HDY
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003650 | | Descriptor: | (2R)-3-(1H-indol-3-yl)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propan-1-ol, (2S)-3-(1H-indol-3-yl)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]propan-1-ol, Non-structural protein 3, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
2WZS
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhu, Y, Suits, M.D.L, Thompson, A, Chavan, S, Dinev, Z, Dumon, C, Smith, N, Moremen, K.W, Xiang, Y, Siriwardena, A, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
3MZO
 
 | |
4G2A
 
 | |
4KTC
 
 | | NS3/NS4A protease with inhibitor | | Descriptor: | (2R,6S,13aR,14aR,16aS)-6-{[(cyclopentyloxy)carbonyl]amino}-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxooctadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 3,4-dihydroisoquinoline-2(1H)-carboxylate, NS4A peptide, Serine protease NS3, ... | | Authors: | Zhang, H, Ballard, J, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Danoprevir (ITMN-191/R7227), a Highly Selective and Potent Inhibitor of Hepatitis C Virus (HCV) NS3/4A Protease.
J.Med.Chem., 57, 2014
|
|
4GH2
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 3-(3-methoxy-4-nitrophenyl)-6-[2-(morpholin-4-yl)ethoxy]-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4AIU
 
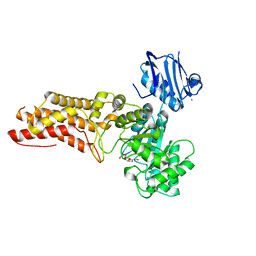 | | A complex structure of BtGH84 | | Descriptor: | (3AR,5R,6S,7R,7AR)-2,5-BIS(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
1H0Z
 
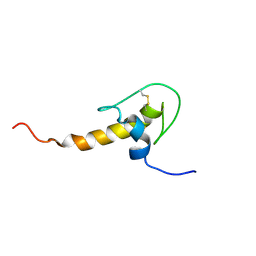 | | LEKTI domain six | | Descriptor: | SERINE PROTEASE INHIBITOR KAZAL-TYPE 5, CONTAINS HEMOFILTRATE PEPTIDE HF6478, HEMOFILTRATE PEPTIDE HF7665 | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
6HW7
 
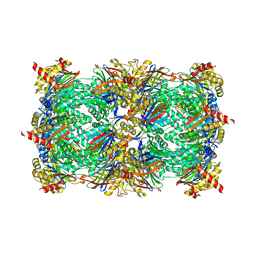 | | Yeast 20S proteasome in complex with 29 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
2X1D
 
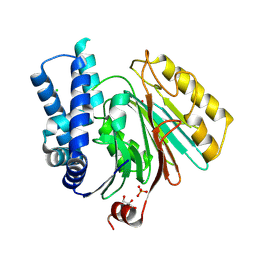 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, ACETATE ION, ACYL-COENZYME, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
4NF6
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and PPDA | | Descriptor: | (2S,3R)-1-(phenanthren-2-ylcarbonyl)piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Jespersen, A, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
4AEI
 
 | | Crystal structure of the AaHII-Fab4C1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALPHA-MAMMAL TOXIN AAH2, CHLORIDE ION, ... | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
3N2K
 
 | | TUBULIN-NSC 613862: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Barbier, P, Dorleans, A, Devred, F, Sanz, L, Allegro, D, Alfonso, C, Knossow, M, Peyrot, V, Andreu, J.M. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers.
J.Biol.Chem., 285, 2010
|
|
4FSW
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 8-chloro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
