4YGT
 
 | |
4SGA
 
 | |
7GGQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-92e193ae-1 (Mpro-x12692) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5SQW
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5014193706 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)-2-hydroxybenzamido]-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SR1
 
 | |
7GI8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIC-UNK-45817b9b-1 (Mpro-P0060) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6FZO
 
 | | SMURFP-Y56F mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Janowski, R, Fuenzalida-Wernera, J.P, Mishra, K, Vetschera, P, Weidenfeld, I, Richter, K, Niessing, D, Ntziachristos, V, Stiel, A.C. | | Deposit date: | 2018-03-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a biliverdin-bound phycobiliprotein: Interdependence of oligomerization and chromophorylation.
J. Struct. Biol., 204, 2018
|
|
4D2P
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 7-({4-[(3-hydroxy-5-methoxyphenyl)amino]benzoyl}amino)-1,2,3,4-tetrahydroisoquinolinium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4TYO
 
 | | PPIase in complex with a non-phosphate small molecule inhibitor. | | Descriptor: | 3-(6-fluoro-1H-benzimidazol-2-yl)-N-(naphthalen-2-ylcarbonyl)-D-alanine, GLYCEROL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Greasley, S.E, Ferre, R.A. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6N9K
 
 | | Beta-lactamase from Escherichia coli str. Sakai | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Beta-lactamase from Escherichia coli str. Sakai
to be published
|
|
7GKH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-UNI-8d415491-1 (Mpro-P0884) | | Descriptor: | (4R)-6,7-dichloro-N-(4-cyclopropylpyridin-3-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4TWI
 
 | | The structure of Sir2Af1 bound to a succinylated histone peptide | | Descriptor: | GLYCEROL, NAD-dependent protein deacylase 1, Succinylated H4 Peptide (aa8-20), ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
4TWK
 
 | | Crystal structure of human two pore domain potassium ion channel TREK1 (K2P2.1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Octyl Glucose Neopentyl Glycol, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Tessitore, A, Goubin, S, Strain-Damerell, C, Mukhopadhyay, S, Kupinska, K, Wang, D, Chalk, R, Berridge, G, Grieben, M, Shrestha, L, Ang, J.H, Mackenzie, A, Quigley, A, Bushell, S.R, Shintre, C.A, Faust, B, Chu, A, Dong, L, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human two pore domain potassium ion channel TREK1 (K2P2.1)
To Be Published
|
|
5C7Q
 
 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
5A3B
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, ALANINE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
4CRC
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | (2S)-2-[[(E)-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]amino]-3-phenyl-N-[4-(1H-1,2,3,4-tetrazol-5-yl)phenyl]propanamide, COAGULATION FACTOR XI, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
6KCC
 
 | |
5SOI
 
 | |
4QNM
 
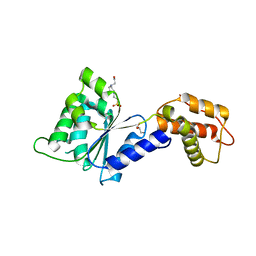 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
2YBV
 
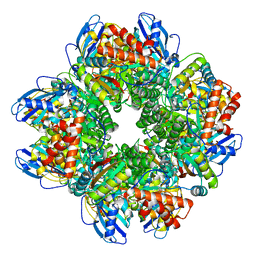 | | STRUCTURE OF RUBISCO FROM THERMOSYNECHOCOCCUS ELONGATUS | | Descriptor: | CHLORIDE ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL SUBUNIT | | Authors: | Terlecka, B, Wilhelmi, V, Bialek, W, Gubernator, B, Szczepaniak, A, Hofmann, E. | | Deposit date: | 2011-03-10 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase from Thermosynechococcus Elongatus
To be Published
|
|
5XNA
 
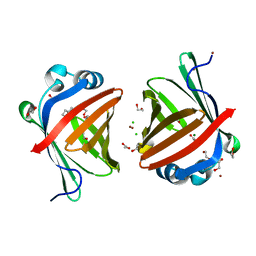 | | Crystal structure of a secretary abundant heat soluble (SAHS) protein from Ramazzottius varieornatus (from dimer sample) | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Fukuda, Y, Miura, Y, Mizohata, E, Inoue, T. | | Deposit date: | 2017-05-19 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural insights into a secretory abundant heat-soluble protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus
FEBS Lett., 591, 2017
|
|
7GH2
 
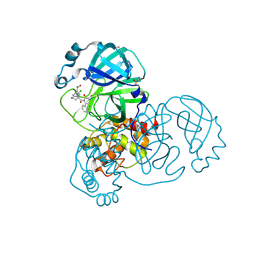 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6CMJ
 
 | | Human CAMKK2 with GSK650393 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropyl)-4-(5-phenyl-1H-pyrrolo[2,3-b]pyridin-3-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Williams, S.P, Reid, R.A, Price, D.J, Drewry, D.H. | | Deposit date: | 2018-03-05 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orally available, brain-penetrant CAMKK2 inhibitor reduces food intake in rodent model.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7GFO
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ROB-IMP-e811baff-1 (Mpro-x11798) | | Descriptor: | 2-(1H-benzotriazol-1-yl)-N-[4-(methylcarbamamido)phenyl]-N-[(thiophen-3-yl)methyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GG3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with AAR-RCN-748c104b-1 (Mpro-x12080) | | Descriptor: | (E)-1-(4,6-dimethoxypyrimidin-2-yl)methanimine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
