8FJ4
 
 | | LSD1-CoREST in complex with T108, short soaking | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-phenylbenzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8F2Z
 
 | | LSD1-CoREST in complex with AW2, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-([1,1'-biphenyl]-4-yl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-09 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8F30
 
 | | LSD1-CoREST in complex with AW2, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-{5-[3-([1,1'-biphenyl]-4-yl)propanoyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-09 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8F59
 
 | | LSD1-CoREST in complex with AW2 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-12 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8FJ7
 
 | | LSD1-CoREST in complex with T108 and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8F6S
 
 | | LSD1-CoREST in complex with T105 | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-(5-methyl-1,3,4-thiadiazol-2-yl)benzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
8FDV
 
 | | LSD1-CoREST in complex N-formyl FAD and SNAG peptide | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, Zinc finger protein SNAI1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-05 | | Release date: | 2024-06-12 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Covalent adduct Grob fragmentation underlies LSD1 demethylase-specific inhibitor mechanism of action and resistance.
Nat Commun, 16, 2025
|
|
4C06
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with MgCl2 | | Descriptor: | MAGNESIUM ION, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
2RG3
 
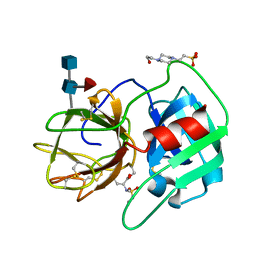 | | Covalent complex structure of elastase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Leukocyte elastase | | Authors: | Huang, W, Yamamoto, Y. | | Deposit date: | 2007-10-02 | | Release date: | 2008-07-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshot of the mechanism of inactivation of human neutrophil elastase by 1,2,5-thiadiazolidin-3-one 1,1-dioxide derivatives.
J.Med.Chem., 51, 2008
|
|
7PFW
 
 | |
7PFX
 
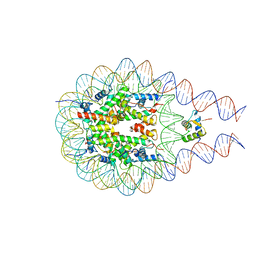 | |
7PFC
 
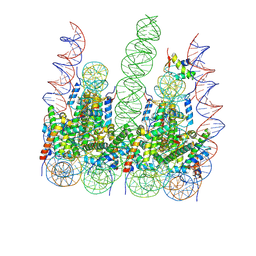 | |
7PET
 
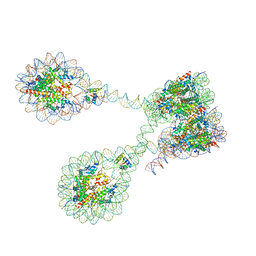 | | The 4x177 nucleosome array containing H1 | | Descriptor: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | Authors: | Dombrowski, M, Cramer, P. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PEX
 
 | |
7PEZ
 
 | |
7PFU
 
 | |
7PF3
 
 | |
7PFA
 
 | |
4C05
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with SAH | | Descriptor: | PROTEIN ARGININE N-METHYLTRANSFERASE 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
2R64
 
 | | Crystal structure of a 3-aminoindazole compound with CDK2 | | Descriptor: | Cell division protein kinase 2, N-[5-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-1H-INDAZOL-3-YL]-2-(4-PIPERIDIN-1-YLPHENYL)ACETAMIDE | | Authors: | Lee, J, Choi, H, Kim, K.H, Jeong, S, Park, J.W, Baek, C.S, Lee, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and biological evaluation of 3,5-diaminoindazoles as cyclin-dependent kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8UKN
 
 | | APO and AMP-PNP bound cAMP-dependent protein kinase A catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2023-10-14 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic, kinetic, and calorimetric investigation of PKA interactions with L-type calcium channels and Rad GTPase.
J.Biol.Chem., 301, 2024
|
|
4BNC
 
 | | Crystal structure of the DNA-binding domain of human ETV1 complexed with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', HUMAN ETV1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Vollmar, M, Froese, D.S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2013-05-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
8RD4
 
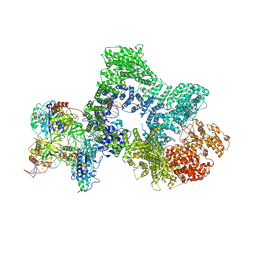 | | Telomeric RAP1:DNA-PK complex | | Descriptor: | DNA (41-MER), DNA-dependent protein kinase catalytic subunit, Telomeric repeat-binding factor 2-interacting protein 1, ... | | Authors: | Eickhoff, P, Fisher, C.E.L, Inian, O, Guettler, S, Douglas, M.E. | | Deposit date: | 2023-12-07 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Chromosome end protection by RAP1-mediated inhibition of DNA-PK.
Nature, 642, 2025
|
|
8W9C
 
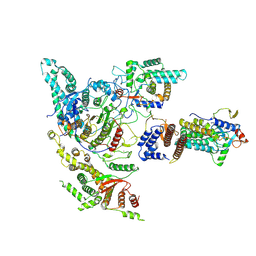 | | Cryo-EM structure of the Rpd3S complex from budding yeast | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, POTASSIUM ION, ... | | Authors: | Wang, C, Zhan, X. | | Deposit date: | 2023-09-05 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures and dynamics of Rpd3S complex bound to nucleosome.
Sci Adv, 10, 2024
|
|
7B5O
 
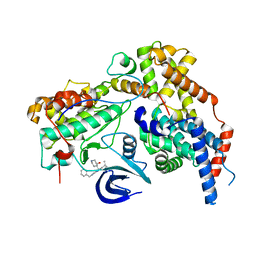 | | Cryo-EM structure of the human CAK bound to ICEC0942 at 2.5 Angstroms resolution | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Greber, B.J, Remis, J, Ali, S, Nogales, E. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.5 angstrom -resolution structure of human CDK-activating kinase bound to the clinical inhibitor ICEC0942.
Biophys.J., 120, 2021
|
|
