5AF9
 
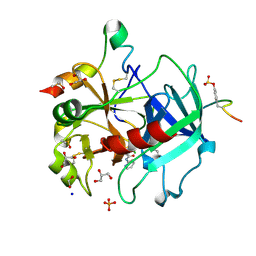 | | Thrombin in complex with 4-Methoxy-N-(2-pyridinyl)benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methoxy-N-(pyridin-2-yl)benzamide, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-01-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Fragments Can Bind Either More Enthalpy or Entropy-Driven: Crystal Structures and Residual Hydration Pattern Suggest Why.
J.Med.Chem., 58, 2015
|
|
4J3V
 
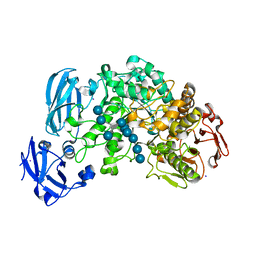 | | Crystal structure of barley limit dextrinase in complex with a branched thio-linked hexasaccharide | | Descriptor: | CALCIUM ION, CHLORIDE ION, IODIDE ION, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
4QIP
 
 | |
7GNL
 
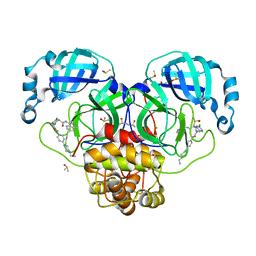 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NIR-WEI-dcc3321b-1 (Mpro-P2757) | | Descriptor: | (4S)-6-chloro-2-{2-[4-(4-ethylpiperazin-1-yl)anilino]-2-oxoethyl}-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4QLV
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 17 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
5SFS
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2c(C)c1nc(nn1c(c2)C)CCc3nc(nn3C)N4CC[C@@H](C4)C, micromolar IC50=0.012855 | | Descriptor: | (4S)-5,8-dimethyl-2-(2-{1-methyl-3-[(3S)-3-methylpyrrolidin-1-yl]-1H-1,2,4-triazol-5-yl}ethyl)[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4QMO
 
 | | MST3 IN COMPLEX WITH Imidazolo-oxindole PKR inhibitor C16 | | Descriptor: | (8Z)-8-(1H-imidazol-5-ylmethylidene)-6,8-dihydro-7H-[1,3]thiazolo[5,4-e]indol-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
2W86
 
 | | Crystal structure of fibrillin-1 domains cbEGF9hyb2cbEGF10, calcium saturated form | | Descriptor: | CALCIUM ION, FIBRILLIN-1, IODIDE ION | | Authors: | Jensen, S.A, Iqbal, S, Lowe, E.D, Redfield, C, Handford, P.A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Interdomain Interactions of a Hybrid Domain: A Disulphide-Rich Module of the Fibrillin/Ltbp Superfamily of Matrix Proteins.
Structure, 17, 2009
|
|
5E28
 
 | | Crystal structure of human carbonic anhydrase II in complex with the 4-(4-aminophenyl)benzenesulfonamide inhibitor | | Descriptor: | 4'-aminobiphenyl-4-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 4-Arylbenzenesulfonamides as Human Carbonic Anhydrase Inhibitors (hCAIs): Synthesis by Pd Nanocatalyst-Mediated Suzuki-Miyaura Reaction, Enzyme Inhibition, and X-ray Crystallographic Studies.
J.Med.Chem., 59, 2016
|
|
4ZU4
 
 | | X-ray structure of the 3,4-ketoisomerase domain of FdtD from Shewanella denitrificans | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
5ABJ
 
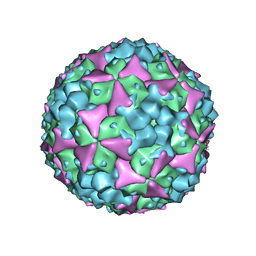 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
5ZJW
 
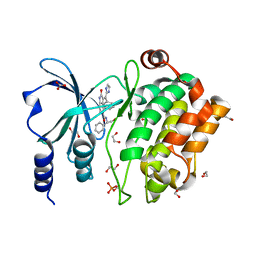 | | Crystal Structure of PAK4 in complex with inhibitor CZg353 | | Descriptor: | (3Z)-N-[(1S)-2-hydroxy-1-phenylethyl]-3-[(1H-imidazol-2-yl)(phenyl)methylidene]-2-oxo-2,3-dihydro-1H-indole-5-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2018-03-22 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal Structure of PAK4 in complex with inhibitor CZg353
To be published
|
|
5AIU
 
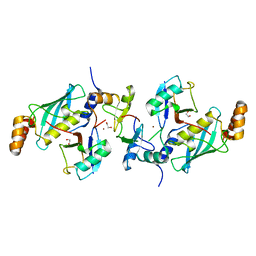 | | A complex of RNF4-RING domain, Ubc13-Ub (isopeptide crosslink) | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF4, POLYUBIQUITIN-C, ... | | Authors: | Branigan, E, Naismith, J.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Ring Catalyzed Synthesis of K63 Linked Ubiquitin Chains
Nat.Struct.Mol.Biol., 22, 2015
|
|
5UX4
 
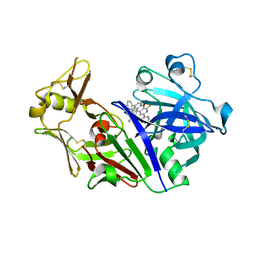 | | Crystal Structure of Rat Cathepsin D with (5S)-3-(5,6-dihydro-2H-pyran-3-yl)-1-fluoro- 7-(2-fluoropyridin-3-yl)spiro[chromeno[2,3- c]pyridine-5,4'-[1,3]oxazol]-2'-amine | | Descriptor: | (5S)-3-(5,6-dihydro-2H-pyran-3-yl)-1-fluoro-7-(2-fluoropyridin-3-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sickmier, A. | | Deposit date: | 2017-02-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthene beta-amyloid cleaving enzyme (BACE) inhibitors with improved selectivity against Cathepsin D.
Medchemcomm, 8, 2017
|
|
5A1A
 
 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | Authors: | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
3IWO
 
 | |
7EHF
 
 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmB1 | | Descriptor: | 1,2-ETHANEDIOL, 16S rRNA methyltransferase, CHLORIDE ION, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Acquired 16S rRNA Methyltransferase NpmB1 Conferring Pan-Aminoglycoside Resistance.
Antimicrob.Agents Chemother., 65, 2021
|
|
5E8R
 
 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | CHLORIDE ION, N-methyl-N-({4-[4-(propan-2-yloxy)phenyl]-1H-pyrrol-3-yl}methyl)ethane-1,2-diamine, Protein arginine N-methyltransferase 6, ... | | Authors: | DONG, A, ZENG, H, LIU, J, TEMPEL, W, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, JIN, J, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases.
Acs Chem.Biol., 11, 2016
|
|
5DKX
 
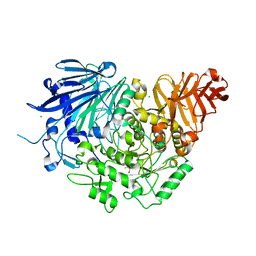 | | Crystal structure of glucosidase II alpha subunit (Tris-bound from) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CHLORIDE ION | | Authors: | Satoh, T, Toshimori, T, Yan, G, Yamaguchi, T, Kato, K. | | Deposit date: | 2015-09-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Sci Rep, 6, 2016
|
|
5DL1
 
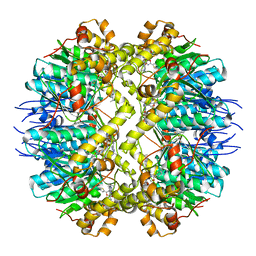 | | ClpP from Staphylococcus aureus in complex with AV145 | | Descriptor: | 1-(propan-2-yl)-N-{[2-(thiophen-2-yl)-1,3-oxazol-4-yl]methyl}-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, ATP-dependent Clp protease proteolytic subunit | | Authors: | Vielberg, M.-T, Groll, M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4QP2
 
 | |
2WR7
 
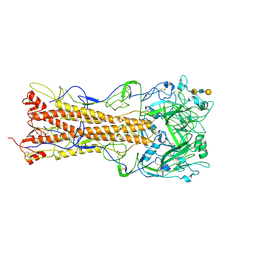 | | the structure of influenza H2 human singapore hemagglutinin with human receptor | | Descriptor: | HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Haire, L.F, Walker, P.A, Coombs, P.J, Russell, R.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2009-08-30 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5EA7
 
 | | Crystal Structure of Inhibitor BMS-433771 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1-cyclopropyl-3-[[1-(4-oxidanylbutyl)benzimidazol-2-yl]methyl]imidazo[4,5-c]pyridin-2-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
2WHL
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
