6TLD
 
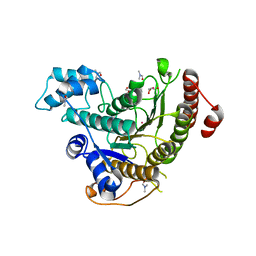 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor 2 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Romier, C. | | Deposit date: | 2019-12-02 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Triazole-Based smHDAC8 Inhibitors.
Chemmedchem, 15, 2020
|
|
9NN2
 
 | | Composite structure of HSV-1 helicase-primase in complex with a forked DNA and amenamevir | | Descriptor: | Amenamevir, DNA (5'-D(P*AP*TP*CP*TP*GP*TP*T)-3'), DNA helicase/primase complex-associated protein, ... | | Authors: | He, Q, Baranovskiy, A.G, Morstadt, L.M, Babayeva, N.D, Lim, C, Tahirov, T.H. | | Deposit date: | 2025-03-04 | | Release date: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of herpesvirus helicase-primase inhibition by pritelivir and amenamevir.
Sci Adv, 11, 2025
|
|
9NNP
 
 | | Composite structure of HSV-1 helicase-primase in complex with a forked DNA | | Descriptor: | DNA (5'-D(P*TP*AP*CP*AP*TP*AP*A)-3'), DNA helicase/primase complex-associated protein, DNA primase, ... | | Authors: | He, Q, Baranovskiy, A.G, Morstadt, L.M, Babayeva, N.D, Lim, C, Tahirov, T.H. | | Deposit date: | 2025-03-05 | | Release date: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of herpesvirus helicase-primase inhibition by pritelivir and amenamevir.
Sci Adv, 11, 2025
|
|
9NQP
 
 | | Composite structure of HSV1 helicase-primase in complex with a forked DNA and pritelivir | | Descriptor: | DNA (5'-D(P*AP*TP*CP*TP*GP*TP*T)-3'), DNA helicase/primase complex-associated protein, DNA primase, ... | | Authors: | He, Q, Baranovskiy, A.G, Morstadt, L.M, Babayeva, N.D, Lim, C, Tahirov, T.H. | | Deposit date: | 2025-03-12 | | Release date: | 2025-11-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of herpesvirus helicase-primase inhibition by pritelivir and amenamevir.
Sci Adv, 11, 2025
|
|
7FPK
 
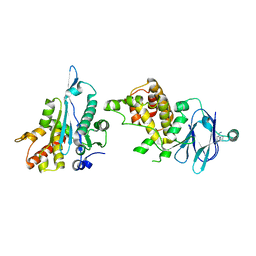 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P10E03 from the F2X-Universal Library | | Descriptor: | 1-[2-(pyrrolidin-1-yl)pyridin-3-yl]methanamine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FKX
 
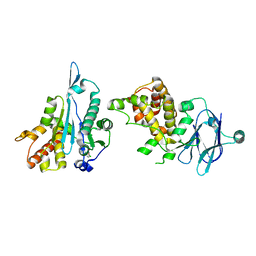 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04G02 from the F2X-Universal Library | | Descriptor: | 1-(4-chlorophenyl)cyclobutane-1-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3HG1
 
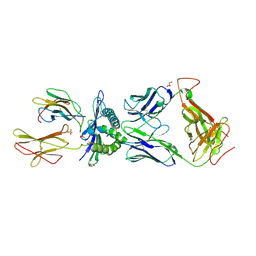 | | Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor | | Descriptor: | Beta-2-microglobulin, CANCER/MART-1, GLYCEROL, ... | | Authors: | Cole, D.K, Yuan, F, Rizkallah, P.J, Miles, J.J, Gostick, E, Price, D.A, Gao, G.F, Jakobsen, B.K, Sewell, A.K. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor.
J.Biol.Chem., 284, 2009
|
|
2AJ3
 
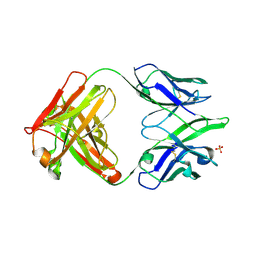 | | Crystal Structure of a Cross-Reactive HIV-1 Neutralizing CD4-Binding Site Antibody Fab m18 | | Descriptor: | Fab m18, Heavy Chain, Light Chain, ... | | Authors: | Prabakaran, P, Gan, J, Wu, Y.Q, Zhang, M.Y, Dimitrov, D.S, Ji, X. | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural mimicry of CD4 by a cross-reactive HIV-1 neutralizing antibody with CDR-H2 and H3 containing unique motifs.
J.Mol.Biol., 357, 2006
|
|
6KL5
 
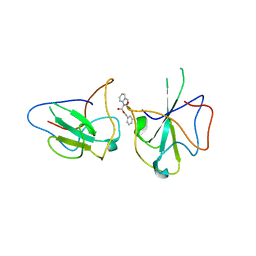 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
9JT5
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S30-1023x | | Descriptor: | 5-[[(4-carboxy-5-methyl-furan-2-yl)methyl-[[(3~{R})-6-ethoxy-3,4-dihydro-2~{H}-chromen-3-yl]carbonyl]amino]methyl]-2-methyl-furan-3-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Jiang, J, Sun, H, Fang, P. | | Deposit date: | 2024-10-02 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S30-1023x
To Be Published
|
|
3QCR
 
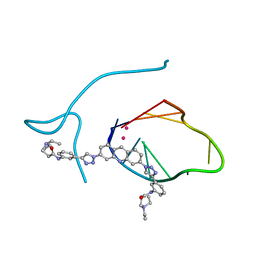 | | Incomplete structural model of a human telomeric DNA quadruplex-acridine complex. | | Descriptor: | Human telomeric repeat deoxyribonucleic acid, N,N'-[acridine-3,6-diylbis(1H-1,2,3-triazole-1,4-diylbenzene-3,1-diyl)]bis[3-(diethylamino)propanamide], POTASSIUM ION | | Authors: | Collie, G.W, Neidle, S, Parkinson, G.N. | | Deposit date: | 2011-01-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of telomeric RNA quadruplex-acridine ligand recognition.
J.Am.Chem.Soc., 133, 2011
|
|
9JT6
 
 | |
4A0D
 
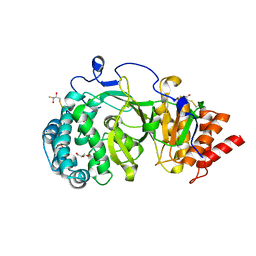 | | Structure of unliganded human PARG catalytic domain | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Overman, R, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
5GAP
 
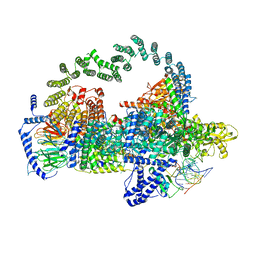 | | Body region of the U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, Pre-mRNA-processing factor 31, Pre-mRNA-splicing factor 6, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Oubridge, C, Bai, X.C, Newman, A, Scheres, S, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
3HD6
 
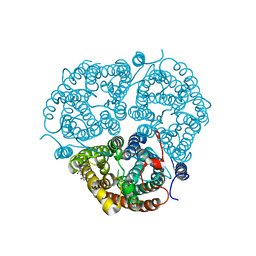 | | Crystal Structure of the Human Rhesus Glycoprotein RhCG | | Descriptor: | Ammonium transporter Rh type C, octyl beta-D-glucopyranoside | | Authors: | Gruswitz, F, Chaudhary, S, Ho, J.D, Pezeshki, B, Ho, C.-M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-05-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of human Rh based on structure of RhCG at 2.1 A.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6A5T
 
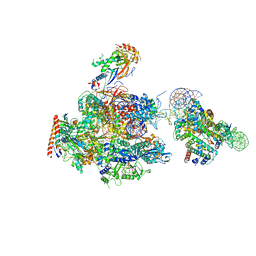 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
5G57
 
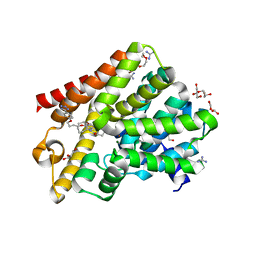 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
1RP4
 
 | | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, CADMIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Gross, E, Kastner, D.B, Kaiser, C.A, Fass, D. | | Deposit date: | 2003-12-03 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of ero1p, source of disulfide bonds for oxidative protein folding in the cell.
Cell(Cambridge,Mass.), 117, 2004
|
|
6TTO
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FORMIC ACID, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
3LXK
 
 | | Structural and Thermodynamic Characterization of the TYK2 and JAK3 Kinase Domains in Complex with CP-690550 and CMP-6 | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Tyrosine-protein kinase JAK3 | | Authors: | Chrencik, J.E, Patny, A, Leung, I.K, Korniski, B, Emmons, T.L, Benson, T.E. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6.
J.Mol.Biol., 400, 2010
|
|
9HD5
 
 | | Crystal structure of CD73 (ecto-5'-nucleotidase) in complex with the AOPCP derivative PSB19427 in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[2-chloranyl-6-[[4-[1-(2-fluoranylethyl)-1,2,3-triazol-4-yl]phenyl]methyl-propyl-amino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N, Moschuetz, S, Dobelmann, C, Schmies, C.C, Jacobson, K.A, Muller, C.E, Junker, A. | | Deposit date: | 2024-11-11 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Positron emission tomography (PET) tracer enables imaging of CD73 expression in cancer
To Be Published
|
|
5DJV
 
 | |
4AVB
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AVP
 
 | | Crystal structure of the DNA-binding domain of human ETV1. | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Krojer, T, Chaikuad, A, Filippakopoulos, P, Canning, P, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-28 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
1RBU
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
