2J5K
 
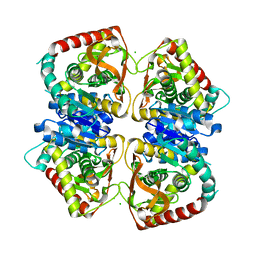 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
3C7E
 
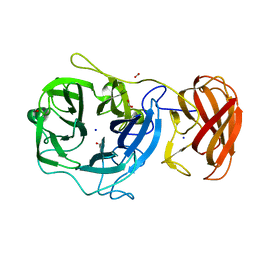 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7G
 
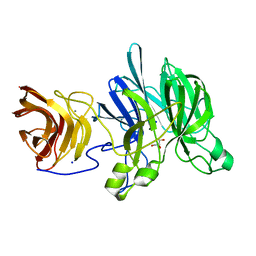 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
4OIF
 
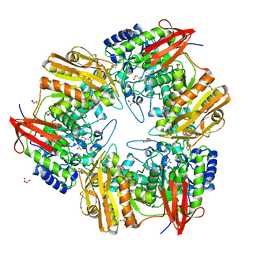 | | 3D structure of Gan42B, a GH42 beta-galactosidase from G. | | Descriptor: | Beta-galactosidase, GLYCEROL, ZINC ION | | Authors: | Solomon, H.V, Tabachnikov, O, Feinberg, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of GanB, a GH42 intracellular beta-galactosidase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3CRW
 
 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
6MYR
 
 | | Avian mitochondrial complex II with thiapronil bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Huang, L.-S, Luemmen, P, Berry, E.A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
4QDD
 
 | | Crystal structure of 3-ketosteroid-9-alpha-hydroxylase 5 (KshA5) from R. rhodochrous in complex with 1,4-30Q-CoA | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
6MYS
 
 | | Avian mitochondrial complex II with Atpenin A5 bound, sidechain outside | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
6MYQ
 
 | | Avian mitochondrial complex II with ferulenol bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
3C7F
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
6MYP
 
 | | Avian mitochondrial complex II with TTFA (thenoyltrifluoroacetone) bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, BICARBONATE ION, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYT
 
 | | Avian mitochondrial complex II with Atpenin A5 bound, sidechain in pocket | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYO
 
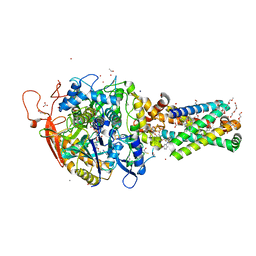 | | Avian mitochondrial complex II with flutolanyl bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6MYU
 
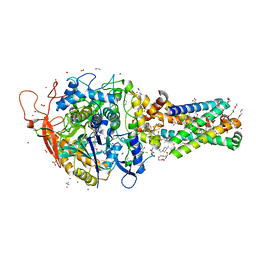 | | Avian mitochondrial complex II crystallized in the presence of HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
3MAN
 
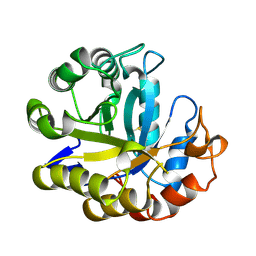 | | MANNOHEXAOSE COMPLEX OF THERMOMONOSPORA FUSCA BETA-MANNANASE | | Descriptor: | PROTEIN (BETA-MANNANASE), beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Hilge, M, Gloor, S.M, Piontek, K. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca - substrate specificity in glycosyl hydrolase family 5.
Structure, 6, 1998
|
|
6NF8
 
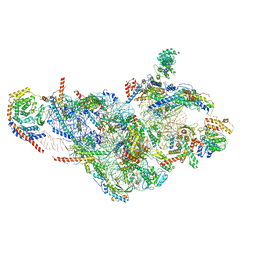 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit -Class I | | Descriptor: | 28S ribosomal RNA, mitochondria, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6NEQ
 
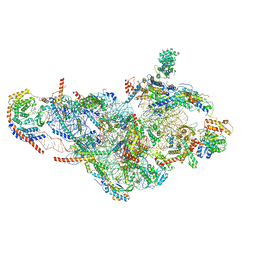 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit-Class-II | | Descriptor: | 28S ribosomal RNA, mitochondrial, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
1NMB
 
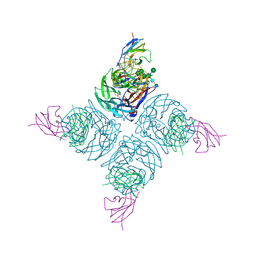 | | THE STRUCTURE OF A COMPLEX BETWEEN THE NC10 ANTIBODY AND INFLUENZA VIRUS NEURAMINIDASE AND COMPARISON WITH THE OVERLAPPING BINDING SITE OF THE NC41 ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB NC10, ... | | Authors: | Malby, R.L, Tulip, W.R, Colman, P.M. | | Deposit date: | 1995-01-17 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody
Structure, 2, 1994
|
|
4B90
 
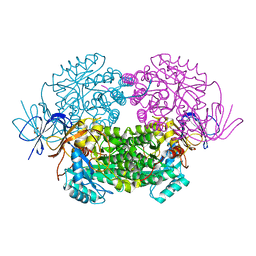 | | Crystal structure of WT human CRMP-5 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-RELATED PROTEIN 5 | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into the Oligomerization of Crmps: Crystal Structure of Human Collapsin Response Mediator Protein 5.
J.Neurochem., 125, 2013
|
|
6N68
 
 | |
4QDF
 
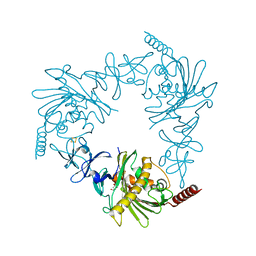 | | Crystal structure of apo KshA5 and KshA1 in complex with 1,4-30Q-CoA from R. rhodochrous | | Descriptor: | 3-ketosteroid 9alpha-hydroxylase oxygenase, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Penfield, J, Worrall, L.J, Strynadka, N.C, Eltis, L.D. | | Deposit date: | 2014-05-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Substrate specificities and conformational flexibility of 3-ketosteroid 9 alpha-hydroxylases.
J.Biol.Chem., 289, 2014
|
|
7QO1
 
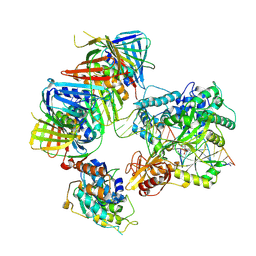 | | complex of DNA ligase I and FEN1 on PCNA and DNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, Flap endonuclease 1, ... | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
2N9X
 
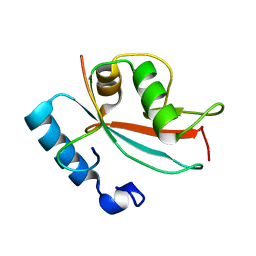 | | LC3 FUNDC1 complex structure | | Descriptor: | FUN14 domain-containing protein 1, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Xia, B, Kuang, Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the phosphorylation of FUNDC1 LIR as a molecular switch of mitophagy.
Autophagy, 12, 2016
|
|
