3FI4
 
 | | P38 kinase crystal structure in complex with RO4499 | | Descriptor: | (2S)-1-{[3-(2-chlorophenyl)-6-(2,4-difluorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-yl]amino}propan-2-ol, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Knapp, M, Dunten, P. | | Deposit date: | 2008-12-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mapping Binding Pocket Volume: Potential Applications towards Ligand Design and Selectivity
To be Published
|
|
3FLS
 
 | |
5L3B
 
 | | Human LSD1/CoREST: LSD1 D556G mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
8KE0
 
 | | Structure of H1.2 bound to the nucleosome | | Descriptor: | DNA (193-MER), Histone H1.2, Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Echigoya, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-08-11 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into how DEK nucleosome binding facilitates H3K27 trimethylation in chromatin.
Nat.Struct.Mol.Biol., 2025
|
|
3FSF
 
 | | P38 kinase crystal structure in complex with 3-(2,6-Dichloro-phenyl)-7-[4-(2-diethylamino-ethoxy)-phenylamino]-1-methyl-3,4-dihydro-1H-pyrimido[4,5-d]pyrimidin-2-one | | Descriptor: | 3-(2,6-dichlorophenyl)-7-({4-[(diethylamino)methoxy]phenyl}amino)-1-methyl-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Bertrand, J, Lovejoy, B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Discovery of Pamapimod and R1487 as Orally Bioavailable and Highly Selective Inhibitors of p38 Map Kinase
To be Published
|
|
3FSK
 
 | | P38 kinase crystal structure in complex with RO6257 | | Descriptor: | 3-(2-chlorophenyl)-7-[(trans-4-hydroxycyclohexyl)amino]-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Bertrand, J, Takahara, P, Villasenor, A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Binding Pocket Volume: Potential Applications towards Ligand Design and Selectivity
To be Published
|
|
1XJL
 
 | |
4FTG
 
 | |
6M95
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping: compound 1 | | Descriptor: | (4-benzylpiperidin-1-yl)[2-methoxy-4-(methylsulfanyl)phenyl]methanone, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
6QTC
 
 | | tSH2 domain of transcription elongation factor Spt6 complexed with tyrosine phosphorylated CTD | | Descriptor: | Tyrosine phosphorylated CTD, tSH2 domain of transcription elongation factor Spt6 | | Authors: | Brazda, P, Krejcikova, M, Smirakova, E, Kubicek, K, Stefl, R. | | Deposit date: | 2019-02-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | tSH2 domain of transcription elongation factor Spt6 complexed with tyrosine phosphorylated CTD
To Be Published
|
|
8JAL
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6UXW
 
 | | SWI/SNF nucleosome complex with ADP-BeFx | | Descriptor: | 601 sequence bottom strand, 601 sequence top strand, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | He, Y, Han, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (8.96 Å) | | Cite: | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
6R6H
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
7JOA
 
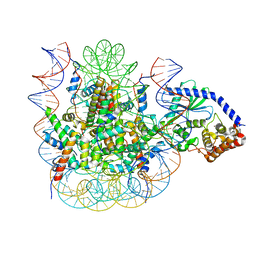 | | 2:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
4FJC
 
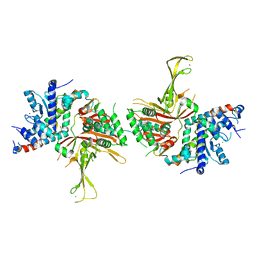 | | Structure of the SAGA Ubp8/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | GLYCEROL, Protein SUS1, SAGA-associated factor 11, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-11 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
4FK5
 
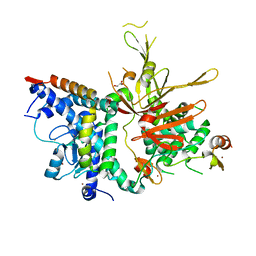 | | Structure of the SAGA Ubp8(S144N)/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
7JO9
 
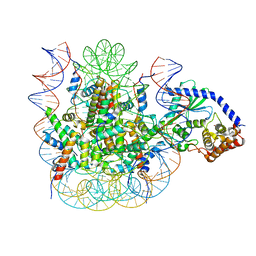 | | 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
6OHD
 
 | | P38 in complex with T-3220137 | | Descriptor: | 3-(3-tert-butyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)-4-methyl-N-(1,2-oxazol-3-yl)benzamide, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]Pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 2.
Chemmedchem, 14, 2019
|
|
5MPA
 
 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
9EOZ
 
 | | Human OGG1 bound to a nucleosome core particle with 8-oxodGuo lesion at SHL6.0 | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.3, ... | | Authors: | Ren, M, Gut, F, Fan, Y, Hopfner, K.-P, Zhou, C. | | Deposit date: | 2024-03-16 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for human OGG1 processing 8-oxodGuo within nucleosome core particles.
Nat Commun, 15, 2024
|
|
5ML5
 
 | | Human p38alpha MAPK in complex with imidazolyl pyridine inhibitor 11b | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-methylsulfanyl-1~{H}-imidazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
5MP9
 
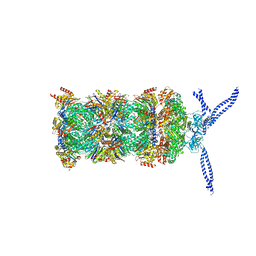 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TXP
 
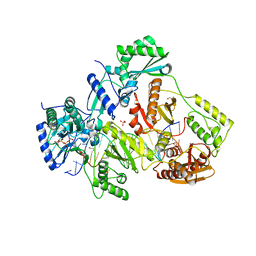 | | STRUCTURE OF Q151M complex (A62V, V75I, F77L, F116Y, Q151M) mutant HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
4FIP
 
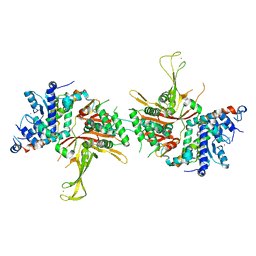 | | Structure of the SAGA Ubp8(S144N)/Sgf11(1-72, Delta-ZnF)/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Ringel, A.E, Wolberger, C. | | Deposit date: | 2012-06-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | A Role for Intersubunit Interactions in Maintaining SAGA Deubiquitinating Module Structure and Activity.
Structure, 20, 2012
|
|
