1MA7
 
 | |
8H3Q
 
 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | Descriptor: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8GQ6
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8H33
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.86 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H35
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.41 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H34
 
 | | Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.99 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3R
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4F52
 
 | | Structure of a Glomulin-RBX1-CUL1 complex | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
6NDL
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with a sulfonamide inhibitor | | Descriptor: | 1-[4-(6-aminopurin-9-yl)butylsulfamoyl]-3-[4-[(4~{S})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butyl]urea, Biotin Protein Ligase, GLYCEROL | | Authors: | Marshall, A.C, Polyak, S.W, Bruning, J.B, Lee, K. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfonamide-Based Inhibitors of Biotin Protein Ligase as New Antibiotic Leads.
Acs Chem.Biol., 14, 2019
|
|
7V8F
 
 | | Crystal structure of UBE2L3 bound to HOIP RING1 domain. | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin-conjugating enzyme E2 L3, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z8R
 
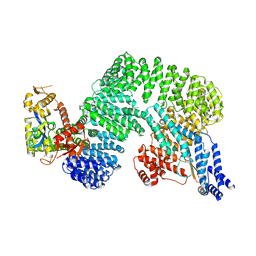 | | CAND1-CUL1-RBX1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8E40
 
 | | Full-length APOBEC3G in complex with HIV-1 Vif, CBF-beta, and fork RNA | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, RNA, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Liu, S, Yang, H, Shiriaeva, A, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase.
Sci Adv, 9, 2023
|
|
1E8C
 
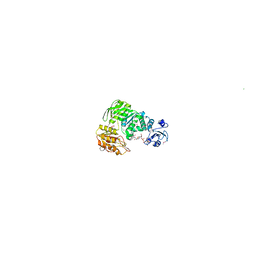 | | Structure of MurE the UDP-N-acetylmuramyl tripeptide synthetase from E. coli | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CHLORIDE ION, UDP-N-ACETYLMURAMOYLALANYL-D-GLUTAMATE--2,6-DIAMINOPIMELATE LIGASE, ... | | Authors: | Gordon, E.J, Chantala, L, Dideberg, O. | | Deposit date: | 2000-09-19 | | Release date: | 2001-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
3CRX
 
 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
6FE8
 
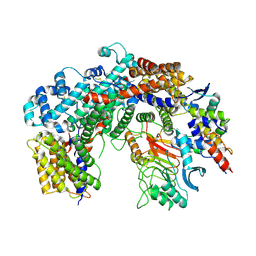 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | Deposit date: | 2017-12-30 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
7XZZ
 
 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
2LGV
 
 | | Rbx1 | | Descriptor: | E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Spratt, D.E, Shaw, G.S. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective recruitment of an e2~ubiquitin complex by an e3 ubiquitin ligase.
J.Biol.Chem., 287, 2012
|
|
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
2B5N
 
 | | Crystal Structure of the DDB1 BPB Domain | | Descriptor: | ISOPROPYL ALCOHOL, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
8RAS
 
 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8RDJ
 
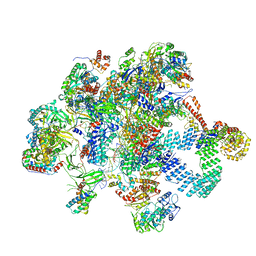 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8OL1
 
 | |
