1UY1
 
 | |
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MKM
 
 | |
6GJA
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - H229A MUTANT | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
7MKL
 
 | |
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
6GQ4
 
 | | Neisseria gonorrhoeae Adhesin Complex Protein | | Descriptor: | Adhesin, SODIUM ION | | Authors: | Orr, C.M, Tews, I. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the RecombinantNeisseria gonorrhoeaeAdhesin Complex Protein (rNg-ACP) and Generation of Murine Antibodies with Bactericidal Activity against Gonococci.
mSphere, 3, 2018
|
|
1TM7
 
 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59Y mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-10 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
6MRY
 
 | | NoD173 plant defensin | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Caria, S, Kvansakul, M. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional characterization of the membrane-permeabilizing activity ofNicotiana occidentalisdefensin NoD173 and protein engineering to enhance oncolysis.
Faseb J., 33, 2019
|
|
1SVE
 
 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 1 | | Descriptor: | (4R)-4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOIC ACID (3R)-3-[(PYRIDINE-4-CARBONYL)AMINO]-AZEPAN-4-YL ESTER, N-OCTANOYL-N-METHYLGLUCAMINE, SODIUM ION, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
7K9A
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-hydroxy-N-[(1R)-2-{5-[(4-{[2-(hydroxymethyl)-1H-imidazol-1-yl]methyl}phenyl)ethynyl]-1H-benzotriazol-1-yl}-1-(methylsulfanyl)ethyl]formamide, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
6GN6
 
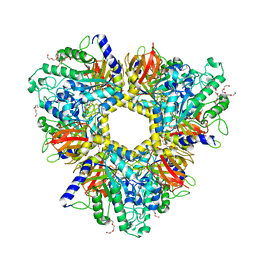 | | Alpha-L-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus | | Descriptor: | Alpha-L-fucosidase, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Kovalova, T, Koval, T, Lipovova, P, Dohnalek, J. | | Deposit date: | 2018-05-30 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active site complementation and hexameric arrangement in the GH family 29; a structure-function study of alpha-l-fucosidase isoenzyme 1 from Paenibacillus thiaminolyticus.
Glycobiology, 29, 2019
|
|
6MW5
 
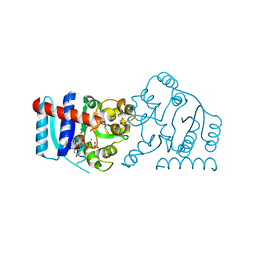 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Platinum | | Descriptor: | 1,2-ETHANEDIOL, PLATINUM (II) ION, UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
6MX8
 
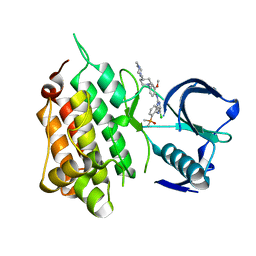 | | Crystal structure of anaplastic lymphoma kinase (ALK) bound by Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, ALK tyrosine kinase receptor | | Authors: | Dougan, D.R, Zhou, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Brigatinib (AP26113), a Phosphine Oxide-Containing, Potent, Orally Active Inhibitor of Anaplastic Lymphoma Kinase.
J. Med. Chem., 59, 2016
|
|
6GG4
 
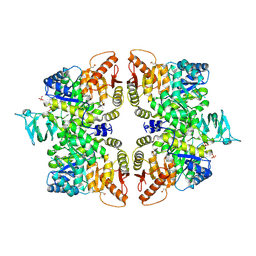 | | Crystal structure of M2 PYK in complex with Phenyalanine. | | Descriptor: | PHENYLALANINE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | McNae, I.W, Yuan, M, Walkinshaw, M.D. | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor.
Biochem. J., 475, 2018
|
|
6N0Q
 
 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
6N6L
 
 | | Crystal Structure of ATPase delta 1-79 Spa47 R189A R191A mutant | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N74
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R271E | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N6Z
 
 | |
6N81
 
 | |
1ULA
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
6MOO
 
 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MW8
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
