5LXQ
 
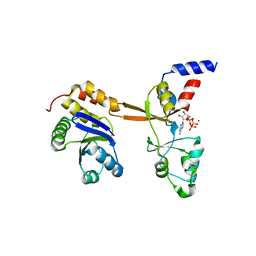 | | Structure of PRL-1 in complex with the Bateman domain of CNNM2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Metal transporter CNNM2, Protein tyrosine phosphatase type IVA 1, ... | | Authors: | GIMENEZ-Mascarell, P, Oyenarte, I, Hardy, S, Breiderhoff, T, Stuiver, M, Kostantin, E, Diercks, T, Pey, A.L, Ereno-ORBEA, J, Martinez-Chantar, M.L, Khalaf-Nazzal, R, Claverie-Martin, F, Muller, D, Tremblay, M, Martinez-Cruz, L.A. | | Deposit date: | 2016-09-22 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | Structural Basis of the Oncogenic Interaction of Phosphatase PRL-1 with the Magnesium Transporter CNNM2.
J. Biol. Chem., 292, 2017
|
|
5LZA
 
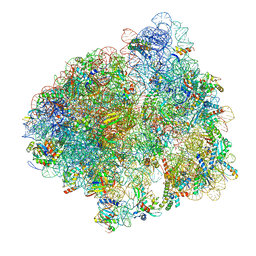 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
1DSC
 
 | |
1DY8
 
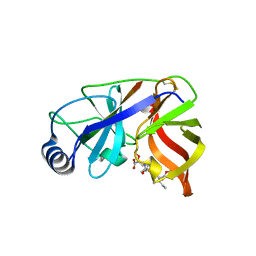 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
9GGV
 
 | | Human KRas4A (GDP) in complex with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(7-chloranyl-3-ethyl-4-oxidanyl-1,3-benzothiazol-2-yl)sulfamoyl]-~{N}-(2-hydroxyethyl)-2-oxidanyl-~{N}-piperidin-4-yl-benzamide, GTPase KRas, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
5LY6
 
 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
1DWN
 
 | | Structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7 A resolution | | Descriptor: | PHAGE COAT PROTEIN | | Authors: | Tars, K, Fridborg, K, Bundule, M, Liljas, L. | | Deposit date: | 1999-12-09 | | Release date: | 2000-02-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure Determination of Phage Pp7 from Pseudomonas Aeruginosa: From Poor Data to a Good Map
Acta Crystallogr.,Sect.D, 56, 2000
|
|
9GH1
 
 | | Human KRas4A (GMPPNP) in complex with compound 34 | | Descriptor: | (3Z)-18-(4-aminocyclohexyl)-21-[[4-(2-aminoethylamino)cyclohexyl]amino]-7-chloro-10-hydroxy-2,2-dioxo-2lambda6,5-dithia-3,12,18,22-tetrazatetracyclo[18.3.1.04,12.06,11]tetracosa-1(23),3,6,8,10,20(24),21-heptaen-19-one, COBALT (II) ION, GTPase KRas, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
1DXX
 
 | |
1DY5
 
 | | Deamidated derivative of bovine pancreatic ribonuclease | | Descriptor: | ACETATE ION, ISOPROPYL ALCOHOL, RIBONUCLEASE A, ... | | Authors: | Esposito, L, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2000-01-27 | | Release date: | 2000-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The Ultrahigh Resolution Crystal Structure of Ribonuclease A Containing an Isoaspartyl Residue: Hydration and Sterochemical Analysis.
J.Mol.Biol., 297, 2000
|
|
5M0O
 
 | | Crystal structure of cytochrome P450 OleT H85Q in complex with arachidonic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
9G4B
 
 | | Human KRas4A (GDP) in complex with compound 15 | | Descriptor: | (3Z)-7-chloro-10,21-dihydroxy-2,2-dioxo-18-(4-piperidyl)-2-lambda-6,5-dithia-3,12,18-triazatetracyclo[18.3.1.04,12.06,11]tetracosa-1(23),3,6,8,10,20(24),21-heptaen-19-one, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-07-15 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
5LYQ
 
 | | Crystal structure of the Retinoic Acid Receptor alpha in complex with a synthetic spiroketal agonist and a fragment of the TIF2 co-activator. | | Descriptor: | (2~{R})-6,6,9,9-tetramethylspiro[3,4,7,8-tetrahydrobenzo[g]chromene-2,2'-3,4-dihydrochromene]-6'-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Ottmann, C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Designed Spiroketal Protein Modulation.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8PFI
 
 | | Crystal structure of human TLR8 in complex with compound 34 | | Descriptor: | (3~{S})-~{N}-[4-[[5-(1,6-dimethylpyrazolo[3,4-b]pyridin-4-yl)-3-methyl-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-1-yl]methyl]-1-bicyclo[2.2.2]octanyl]morpholine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Discovery of the TLR7/8 Antagonist MHV370 for Treatment of Systemic Autoimmune Diseases.
Acs Med.Chem.Lett., 14, 2023
|
|
1E0Z
 
 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Schweimer, K, Marg, B, Oesterhelt, D, Roesch, P, Sticht, H. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
5LYY
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 3-[2-(4-fluoranylphenoxy)ethyl]-1,3-diazaspiro[4.5]decane-2,4-dione, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
1DIT
 
 | | COMPLEX OF A DIVALENT INHIBITOR WITH THROMBIN | | Descriptor: | ALPHA-THROMBIN, PEPTIDE INHIBITOR CVS995 | | Authors: | Tulinsky, A, Krishnan, R. | | Deposit date: | 1995-07-20 | | Release date: | 1996-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, structure, and structure-activity relationships of divalent thrombin inhibitors containing an alpha-keto-amide transition-state mimetic.
Protein Sci., 5, 1996
|
|
1M40
 
 | | ULTRA HIGH RESOLUTION CRYSTAL STRUCTURE OF TEM-1 | | Descriptor: | BETA-LACTAMASE TEM, PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, ... | | Authors: | Minasov, G, Wang, X, Shoichet, B.K. | | Deposit date: | 2002-07-01 | | Release date: | 2002-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | An ultrahigh resolution structure of TEM-1 beta-lactamase suggests a role for Glu166 as the general base in acylation.
J.Am.Chem.Soc., 124, 2002
|
|
5LZF
 
 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
1DWU
 
 | | Ribosomal protein L1 | | Descriptor: | RIBOSOMAL PROTEIN L1 | | Authors: | Tishchenko, S.V, Nevskaya, N.A, Pavelyev, M.N, Nikonov, S.V, Garber, M.B, Piendl, W. | | Deposit date: | 1999-12-13 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Ribosomal Protein L1 from Methanococcus Thermolithotrophicus. Functionally Important Structural Invariants on the L1 Surface
Acta Crystallogr.,Sect.D, 58, 2002
|
|
9GGT
 
 | | Human KRas4A (GDP) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
5M0W
 
 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1DZT
 
 | | RMLC FROM SALMONELLA TYPHIMURIUM | | Descriptor: | 3'-O-ACETYLTHYMIDINE-(5' DIPHOSPHATE PHENYL ESTER), 3'-O-ACETYLTHYMIDINE-5'-DIPHOSPHATE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, ... | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
9GH2
 
 | | Human KRas4A (GMPPNP) in complex with compound 36 | | Descriptor: | (3Z)-18-(4-aminocyclohexyl)-21-[4-(4-amino-1-piperidyl)-1-piperidyl]-7-chloro-10-hydroxy-2,2-dioxo-2-lambda-6,5-dithia-3,12,18,22-tetrazatetracyclo[18.3.1.04,12.06,11]tetracosa-1(23),3,6,8,10,20(24),21-heptaen-19-one, COBALT (II) ION, GTPase KRas, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2024-08-14 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Reversible Small Molecule Multivariant Ras Inhibitors Display Tunable Affinity for the Active and Inactive Forms of Ras.
J.Med.Chem., 68, 2025
|
|
5M0Y
 
 | | Crystal Structure of the CohScaA-XDocCipB type II complex from Clostridium thermocellum at 1.5Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cellulosome anchoring protein cohesin region, ... | | Authors: | Pinheiro, B.A, Bras, J.L, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
