2OSC
 
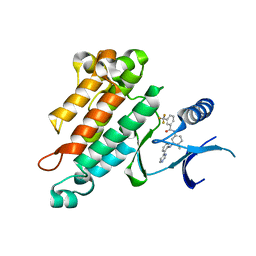 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6SYS
 
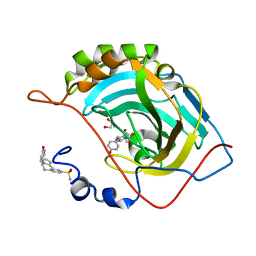 | | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2 | | Descriptor: | (4~{S})-4-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, GLYCEROL, ZINC ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2
To Be Published
|
|
6D5H
 
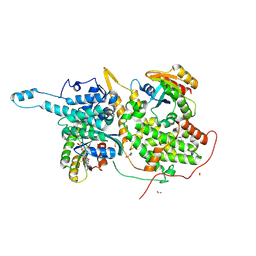 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-4-(2-chlorophenyl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-2-(piperazin-1-yl)-1H-benzimidazole, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
3D8W
 
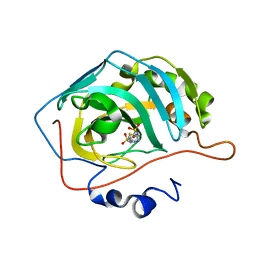 | | Use of a carbonic Anhydrase II, IX Active-site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | 5-[(phenylsulfonyl)amino]-1,3,4-thiadiazole-2-sulfonamide, Carbonic anhydrase II, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-05-26 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
5SGI
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2c(nc1[nH]cnc1c2Oc3cc(ccc3)C)N5CCN(c4ncccc4)CC5, micromolar IC50=0.035 | | Descriptor: | 6-(3-methylphenoxy)-2-[4-(pyridin-2-yl)piperazin-1-yl]-9H-purine, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
1U6V
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
5ISJ
 
 | |
3P4L
 
 | |
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5SDW
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C(Nc1ccc2c(c1)nc([nH]2)c3ccccc3)(=O)c4c(C(N(C)C)=O)cnn4C, micromolar IC50=0.0003757 | | Descriptor: | MAGNESIUM ION, N~4~,N~4~,1-trimethyl-N~5~-(2-phenyl-1H-benzimidazol-5-yl)-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEE
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(ccnn1c2ccccn2)NC(=O)c3nc(ccc3Nc4cncnc4)C5CC5, micromolar IC50=0.003431 | | Descriptor: | 6-cyclopropyl-N-[1-(pyridin-2-yl)-1H-pyrazol-5-yl]-3-[(pyrimidin-5-yl)amino]pyridine-2-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Joseph, C, Rodriguez-Sarmiento, R.M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
6JN4
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor WL-001 | | Descriptor: | ACETATE ION, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
5SED
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(cc2c(c1)nc(nc2N3CCOCC3)OCc4nc(cn4C)c5ccccc5)Cl, micromolar IC50=0.00213 | | Descriptor: | 6-chloro-2-{[(4S)-1-methyl-4-phenyl-4,5-dihydro-1H-imidazol-2-yl]methoxy}-4-(morpholin-4-yl)quinazoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7T67
 
 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
5SEC
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1ccc2c(c1)nc(c(n2)C)CCc3nc(cn3c4ccccc4)c5ccccc5, micromolar IC50=0.000332 | | Descriptor: | 2-[2-(1,4-diphenyl-1H-imidazol-2-yl)ethyl]-3-methylquinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7QTJ
 
 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
3HCC
 
 | | Crystal Structure of hPNMT in Complex With anti-9-amino-5-(trifluromethyl) benzonorbornene and AdoHcy | | Descriptor: | (1S,4R,9S)-5-(trifluoromethyl)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-9-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L, Puri, M. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
5SP1
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC001472868186 | | Descriptor: | 3-{[methyl(pyrido[2,3-b]pyrazin-6-yl)amino]methyl}[1,2,4]triazolo[4,3-a]pyrazin-8(7H)-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SP6
 
 | |
8CBF
 
 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
5ZCL
 
 | | Crystal structure of OsPP2C50 I267L:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
4NR9
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with acetylated lysine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N(6)-ACETYLLYSINE | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
6SWQ
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
1UEJ
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with a substrate, cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CITRIC ACID, Uridine-cytidine kinase 2 | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-05-16 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
6SPO
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
