1DJA
 
 | |
5LLU
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
1DPO
 
 | | STRUCTURE OF RAT TRYPSIN | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Stroud, R.M. | | Deposit date: | 1997-03-31 | | Release date: | 1997-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | 1.59 A structure of trypsin at 120 K: comparison of low temperature and room temperature structures.
Proteins, 10, 1991
|
|
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
1DLO
 
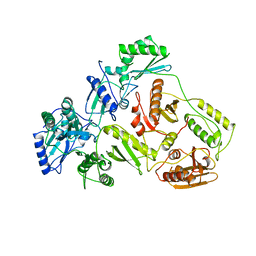 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Ding, J, Das, K, Hughes, S, Arnold, E. | | Deposit date: | 1996-04-17 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms.
Structure, 4, 1996
|
|
1DKG
 
 | |
1DST
 
 | |
5LMS
 
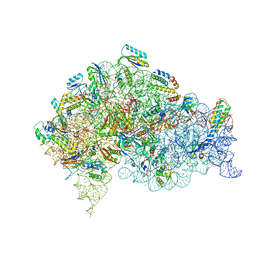 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
1DTC
 
 | |
5LT7
 
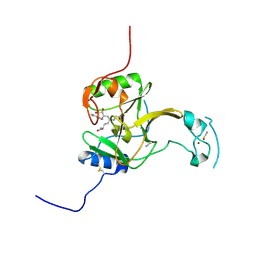 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
1DRN
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GGAGA)R(UGAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*GP*AP*GP*A)-R(P*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-11-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
5LTG
 
 | | Citrate-bound Pichia angusta Atg18 | | Descriptor: | Autophagy-related protein 18, CITRATE ANION, PHOSPHATE ION | | Authors: | Scacioc, A, Kuhnel, K. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Citrate-bound Pichia angusta Atg18
To Be Published
|
|
1DEF
 
 | |
1DPG
 
 | |
5LTT
 
 | |
1DME
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
5LWI
 
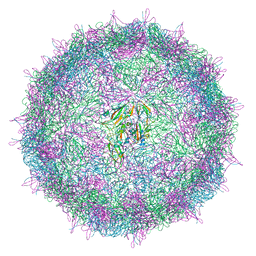 | | Israeli acute paralysis virus heated to 63 degree - empty particle | | Descriptor: | Structural polyprotein, VP1, VP2 | | Authors: | Mullapudi, E, Fuzik, T, Pridal, A, Plevka, P. | | Deposit date: | 2016-09-16 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-electron Microscopy Study of the Genome Release of the Dicistrovirus Israeli Acute Bee Paralysis Virus.
J. Virol., 91, 2017
|
|
1DDF
 
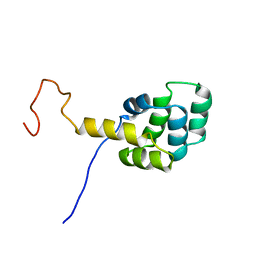 | | FAS DEATH DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FAS | | Authors: | Huang, B, Eberstadt, M, Olejniczak, E, Meadows, R.P, Fesik, S. | | Deposit date: | 1996-11-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain.
Nature, 384, 1996
|
|
1DIP
 
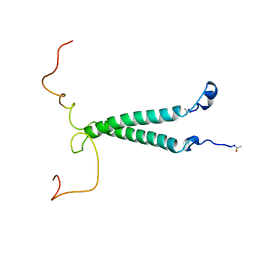 | | THE SOLUTION STRUCTURE OF PORCINE DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE | | Authors: | Roesch, P, Seidel, G, Adermann, K, Schindler, T, Ejchart, A, Jaenicke, R, Forssmann, W.G. | | Deposit date: | 1997-04-09 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide A homolog of the shortsighted gene product.
J.Biol.Chem., 272, 1997
|
|
5LX1
 
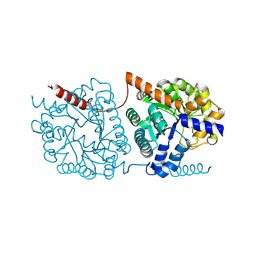 | | Cys-Gly dipeptidase GliJ mutant D304A | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
9GH6
 
 | | CEACAM1 (35-234), focused refinement in the complex with CbpF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Marongiu, G.L, Fink, U, Roderer, D. | | Deposit date: | 2024-08-15 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for immune cell binding of Fusobacterium nucleatum via the trimeric autotransporter adhesin CbpF.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1DEM
 
 | |
1DMD
 
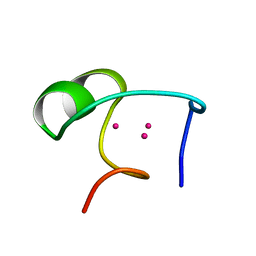 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
5LOP
 
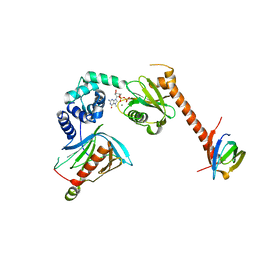 | | Structure of the active form of /K. lactis/ Dcp1-Dcp2-Edc3 decapping complex bound to m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, KLLA0A11308p, KLLA0E01827p, ... | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1DIU
 
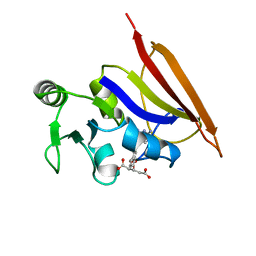 | | DIHYDROFOLATE REDUCTASE (E.C.1.5.1.3) COMPLEX WITH BRODIMOPRIM-4,6-DICARBOXYLATE | | Descriptor: | BRODIMOPRIM-4,6-DICARBOXYLATE, DIHYDROFOLATE REDUCTASE | | Authors: | Morgan, W.D, Birdsall, B, Polshakov, V.I, Sali, D, Kompis, I, Feeney, J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a brodimoprim analogue in its complex with Lactobacillus casei dihydrofolate reductase.
Biochemistry, 34, 1995
|
|
