3LYC
 
 | |
5DHJ
 
 | | PIM1 in complex with Cpd4 (3-methyl-5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridine) | | Descriptor: | 3-methyl-5-(pyridin-3-yl)-2H-pyrazolo[3,4-c]pyridine, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7D5N
 
 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
3DKV
 
 | | Crystal structure of adenylate kinase variant AKlse1 | | Descriptor: | 1,2-ETHANEDIOL, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, Bitto, E.B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of adenylate kinase variant AKlse1.
To be Published
|
|
7YAZ
 
 | | Crystal structure of ZAK in complex with compound YH-186 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-[3-[(3~{S})-1-propanoylpyrrolidin-3-yl]oxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl]-1,2,3-triazol-1-yl]phenyl]-3-phenyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
5GTN
 
 | | Human PPARgamma ligand binding dmain complexed with R35 | | Descriptor: | 2-[4-[5-[(1~{R})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
6EPY
 
 | |
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
7DMN
 
 | | Crystal structure of two pericyclases catalyzing [4+2] cycloaddition | | Descriptor: | Diels-Alderase fsa2, GLYCEROL | | Authors: | Chi, C.B, Wang, Z.D, Liu, T, Zhang, Z.Y, Ma, M. | | Deposit date: | 2020-12-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Fsa2 and Phm7 Catalyzing [4 + 2] Cycloaddition Reactions with Reverse Stereoselectivities in Equisetin and Phomasetin Biosynthesis.
Acs Omega, 6, 2021
|
|
1N1S
 
 | | Trypanosoma rangeli sialidase | | Descriptor: | SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T. cruzi
trans-sialidase
J.Mol.Biol., 325, 2003
|
|
6AKQ
 
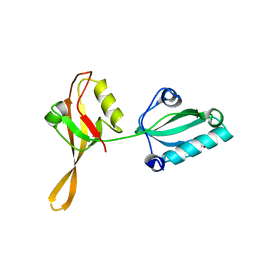 | | The PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 263 and 267 | | Descriptor: | PDZ tandem fragment of A. aeolicus site-2 protease homolog with the PA tag insertion | | Authors: | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
8AIU
 
 | | Mpro of SARS COV-2 in complex with the MG-97 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
3ISD
 
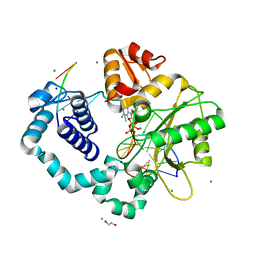 | | Ternary complex of human DNA polymerase beta with an abasic site (THF): DAPCPP mismatch | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, 5'-D(*CP*CP*GP*AP*CP*(3DR)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
1N2M
 
 | | The S53A Proenzyme Structure of Methanococcus jannaschii. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Pyruvoyl-dependent arginine decarboxylase | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-23 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
6SWQ
 
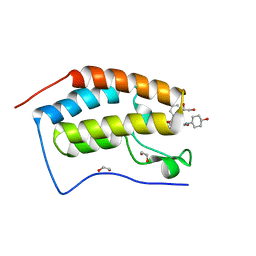 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD2 (GSK046) | | Descriptor: | 1,2-ETHANEDIOL, 4-acetamido-3-fluoranyl-~{N}-(4-oxidanylcyclohexyl)-5-[(1~{S})-1-phenylethoxy]benzamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
3ALC
 
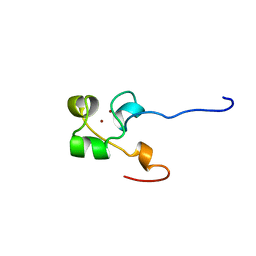 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-03-11 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
8EB2
 
 | |
9L5R
 
 | | Cryo-EM structure of the thermophile spliceosome (state ILS) | | Descriptor: | Anaphase-promoting complex subunit 4-like WD40 domain-containing protein, CCDC12, Cell cycle control protein, ... | | Authors: | Li, Y, Fischer, P, Wang, M, Yuan, R, Meng, W, Luehrmann, R, Lau, B, Hurt, E, Cheng, J. | | Deposit date: | 2024-12-23 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into spliceosome fidelity: DHX35-GPATCH1- mediated rejection of aberrant splicing substrates.
Cell Res., 35, 2025
|
|
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
5D5C
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6KXX
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
5D7P
 
 | | Crystal structure of human Sirt2 in complex with ADPR and EX-243 | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Gerhardt, S, Einsle, O, Jung, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4RE9
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4OOV
 
 | |
7DCE
 
 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
