4MLE
 
 | | Human Glucokinase in Complex with Novel Amino Thiazole Activator | | Descriptor: | 3-(benzyloxy)-N-(4-methyl-1,3-thiazol-2-yl)pyridin-2-amine, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W.C. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a New Class of Glucokinase Activators through Structure-Based Design.
J.Med.Chem., 56, 2013
|
|
7XBA
 
 | | Glutathione S-transferase bound with a covalent inhibitor | | Descriptor: | 3-[3-[[2-[5-[(3,5-dimethyl-4-nitro-pyrazol-1-yl)methyl]furan-2-yl]-5-(methylcarbamoyl)benzimidazol-1-yl]methyl]azetidin-1-yl]sulfonylbenzenesulfonic acid, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Jiang, L.L, Zhou, L. | | Deposit date: | 2022-03-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Glutathione S-transferase bound with a covalent inhibitor
To Be Published
|
|
9FOC
 
 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 11. | | Descriptor: | (2S)-1-[4-[[(3R)-1,1-bis(oxidanylidene)thiolan-3-yl]methyl-methyl-amino]-6-methyl-pyrimidin-2-yl]-N-methyl-pyrrolidine-2-carboxamide, (2S)-1-[4-[[(3S)-1,1-bis(oxidanylidene)thiolan-3-yl]methyl-methyl-amino]-6-methyl-pyrimidin-2-yl]-N-methyl-pyrrolidine-2-carboxamide, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-06-11 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Structural and Molecular Insight into the PWWP1 Domain of NSD2 from the Discovery of Novel Binders Via DNA-Encoded Library Screening.
Acs Med.Chem.Lett., 16, 2025
|
|
3LHU
 
 | | Crystal structure of the mutant I199F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
4MLH
 
 | |
3H69
 
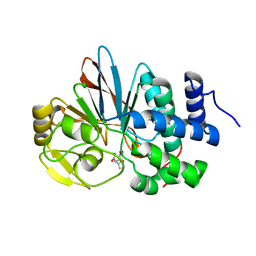 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Zn2+ atoms complexed with endothall | | Descriptor: | (1R,2S,3R,4S)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, Serine/threonine-protein phosphatase 5, ZINC ION | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
2SIM
 
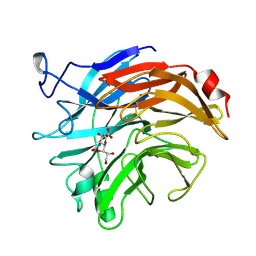 | | THE STRUCTURES OF SALMONELLA TYPHIMURIUM LT2 NEURAMINIDASE AND ITS COMPLEX WITH A TRANSITION STATE ANALOGUE AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SIALIDASE | | Authors: | Taylor, G.L, Crennell, S.J, Garman, E.F, Vimr, E.R, Laver, W.G. | | Deposit date: | 1994-07-15 | | Release date: | 1994-11-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
3LS6
 
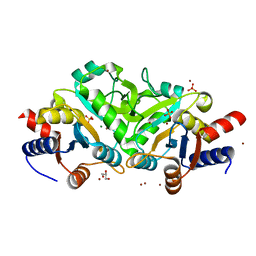 | | Crystal structure of 3,4-Dihydroxy-2-butanone 4-phosphate synthase in complex with sulfate and zinc | | Descriptor: | 3,4-Dihydroxy-2-butanone 4-phosphate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumar, P, Karthikeyan, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Potential anti-bacterial drug target: structural characterization of 3,4-dihydroxy-2-butanone-4-phosphate synthase from Salmonella typhimurium LT2.
Proteins, 78, 2010
|
|
9FDF
 
 | | Human phosphoglycerate kinase in with mixture of products and substrates produced by cross-soaking a TSA crystal | | Descriptor: | 1,3-BISPHOSPHOGLYCERIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cliff, M.J, Waltho, J.P, Bowler, M.W, Baxter, N.J, Bisson, C, Blackburn, G.M. | | Deposit date: | 2024-05-16 | | Release date: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The role of magnesium in catalysis by phosphoglycerate kinase
To be published
|
|
3LLV
 
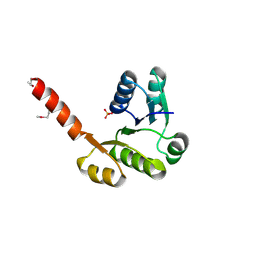 | | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A | | Descriptor: | Exopolyphosphatase-related protein, PHOSPHATE ION | | Authors: | Stein, A.J, Chang, C, Weger, A, Hendricks, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A
To be Published
|
|
5O1W
 
 | |
6GAW
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the complete 55S ribosome. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 16S ribosomal RNA, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
4XSH
 
 | | The complex structure of C3cer exoenzyme and GTP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
5BSK
 
 | | Human HGPRT in complex with (S)-HPEPG, an acyclic nucleoside phosphonate | | Descriptor: | (2-{[(2S)-1-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3-hydroxypropan-2-yl]oxy}ethyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Keough, D.T, Guddat, L.W, Kaiser, M.M, Hockova, D, Wang, T.-H, Janeba, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synthesis and Evaluation of Novel Acyclic Nucleoside Phosphonates as Inhibitors of Plasmodium falciparum and Human 6-Oxopurine Phosphoribosyltransferases.
Chemmedchem, 10, 2015
|
|
4Q7X
 
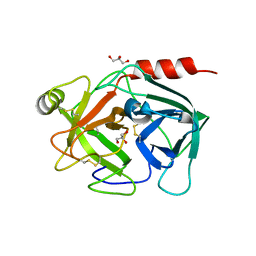 | | Neutrophil serine protease 4 (PRSS57) apo form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Serine protease 57, ... | | Authors: | Eigenbrot, C, Lin, S.J, Dong, K.C. | | Deposit date: | 2014-04-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of neutrophil serine protease 4 reveal an unusual mechanism of substrate recognition by a trypsin-fold protease.
Structure, 22, 2014
|
|
7L3J
 
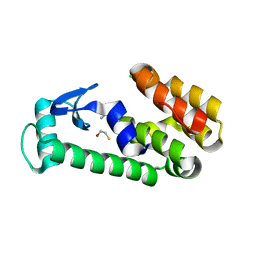 | | T4 Lysozyme L99A - benzylacetate - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L38
 
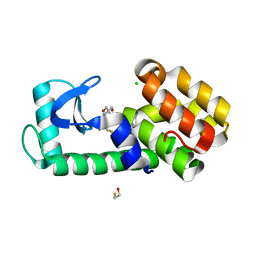 | | T4 Lysozyme L99A - Apo - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
2RI0
 
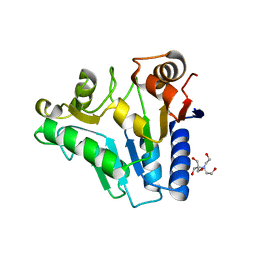 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucosamine-6-phosphate deaminase, SODIUM ION | | Authors: | Li, D, Liu, C, Li, L.F, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
7L37
 
 | | T4 Lysozyme L99A - Apo - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
6PS9
 
 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 17 (5-{2-[(3R)-1-methyl-5-oxopyrrolidin-3-yl]ethyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one) | | Descriptor: | 5-{2-[(3R)-1-methyl-5-oxopyrrolidin-3-yl]ethyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one, Bromodomain-containing protein 4 | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Synthesis and elaboration of N-methylpyrrolidone as an acetamide fragment substitute in bromodomain inhibition.
Bioorg.Med.Chem., 27, 2019
|
|
6IZO
 
 | | Crystal structure of DNA polymerase sliding clamp from Caulobacter crescentus | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER | | Authors: | Jiang, X, Zhang, L, Teng, M, Li, X. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
7L39
 
 | | T4 Lysozyme L99A - toluene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
3HCF
 
 | |
6DR0
 
 | | Class 5 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7L3B
 
 | | T4 Lysozyme L99A - iodobenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
