6TOY
 
 | |
6LCV
 
 | |
6LCU
 
 | |
6Z8L
 
 | | Alpha-Amylase in complex with probe fragments | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.40000367 Å) | | Cite: | Enhancing glycan stability via site-selective fluorination: modulating substrate orientation by molecular design.
Chem Sci, 12, 2020
|
|
7CCS
 
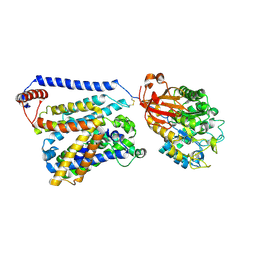 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
7CMH
 
 | | The LAT2-4F2hc complex in complex with tryptophan | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Zhou, J.Y, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex.
Cell Discov, 6, 2020
|
|
7CMI
 
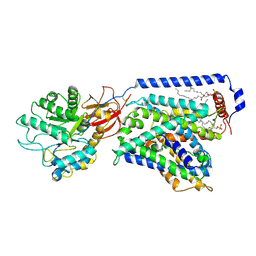 | | The LAT2-4F2hc complex in complex with leucine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Zhou, J.Y, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex.
Cell Discov, 6, 2020
|
|
6YUP
 
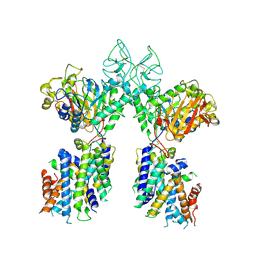 | | Heterotetrameric structure of the rBAT-b(0,+)AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neutral and basic amino acid transport protein rBAT, ... | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YUZ
 
 | | Homodimeric structure of the rBAT complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neutral and basic amino acid transport protein rBAT | | Authors: | Wu, D, Safarian, S, Michel, H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YQC
 
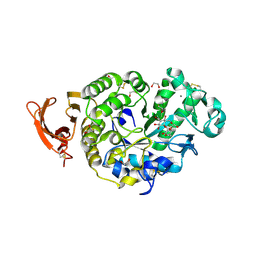 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{R},5~{S},6~{R})-6-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-heptoxy-6-(hydroxymethyl)-3,4-bis(oxidanyl)oxan-2-yl]oxy-5-(hydroxymethyl)cyclohexane-1,2,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQB
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol cyclosulfate inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQ9
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6YQA
 
 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol aziridine inhibitor | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},5~{R})-5-(8-azanyloctylamino)-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
7DSL
 
 | | Overall structure of the LAT1-4F2hc bound with JX-078 | | Descriptor: | (2~{S})-2-azanyl-7-[(2-phenylphenyl)methoxy]-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSQ
 
 | | Overall structure of the LAT1-4F2hc bound with 3,5-diiodo-L-tyrosine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-DIIODOTYROSINE, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSN
 
 | | Overall structure of the LAT1-4F2hc bound with JX-119 | | Descriptor: | (2~{S})-2-azanyl-7-[[2-(1,3-benzoxazol-2-yl)phenyl]methoxy]-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSK
 
 | | Overall structure of the LAT1-4F2hc bound with JX-075 | | Descriptor: | (2~{S})-2-azanyl-7-(naphthalen-1-ylmethoxy)-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
6WNU
 
 | |
6WNI
 
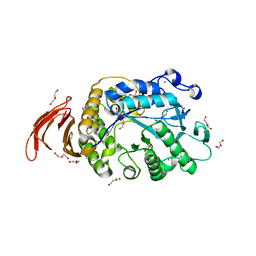 | |
7JJT
 
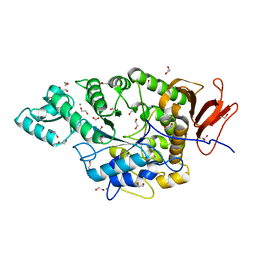 | | Ruminococcus bromii amylase Amy5 (RBR_07800) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-amylase, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The structures of the GH13_36 amylases from Eubacterium rectale and Ruminococcus bromii reveal subsite architectures that favor maltose production
Amylase, 4, 2020
|
|
7LSA
 
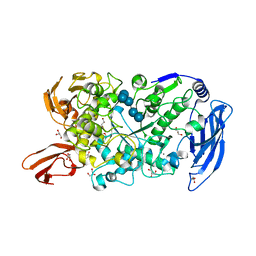 | | Ruminococcus bromii Amy12 with maltoheptaose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LST
 
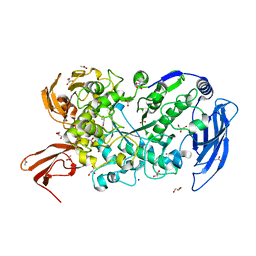 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSU
 
 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSR
 
 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | Descriptor: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7D9C
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-13 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
