5HNZ
 
 | | Structural basis of backwards motion in kinesin-14: plus-end directed nKn669 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
3CHJ
 
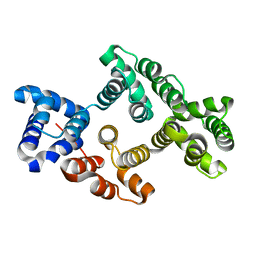 | | Crystal Structure of Alpha-14 Giardin | | Descriptor: | Alpha-14 giardin, CALCIUM ION | | Authors: | Pathuri, P, Luecke, H. | | Deposit date: | 2008-03-10 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Apo and calcium-bound crystal structures of cytoskeletal protein alpha-14 giardin (annexin E1) from the intestinal protozoan parasite Giardia lamblia
J.Mol.Biol., 385, 2009
|
|
3CHK
 
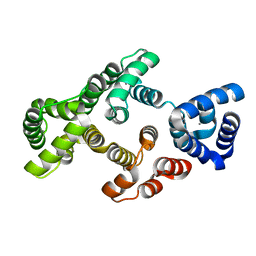 | | Calcium bound structure of alpha-14 giardin | | Descriptor: | Alpha-14 giardin, CALCIUM ION | | Authors: | Pathuri, P, Luecke, H. | | Deposit date: | 2008-03-10 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Apo and calcium-bound crystal structures of cytoskeletal protein alpha-14 giardin (annexin E1) from the intestinal protozoan parasite Giardia lamblia
J.Mol.Biol., 385, 2009
|
|
5HNX
 
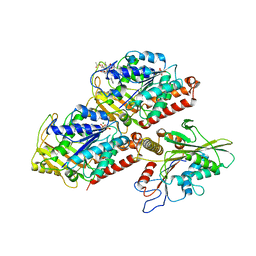 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
3CHL
 
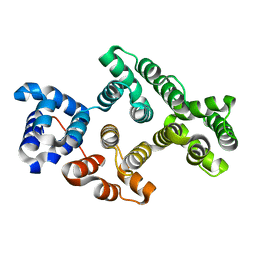 | | Crystal Structure of Alpha-14 Giardin with magnesium bound | | Descriptor: | Alpha-14 giardin, MAGNESIUM ION | | Authors: | Pathuri, P, Luecke, H. | | Deposit date: | 2008-03-10 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Apo and calcium-bound crystal structures of cytoskeletal protein alpha-14 giardin (annexin E1) from the intestinal protozoan parasite Giardia lamblia
J.Mol.Biol., 385, 2009
|
|
5Z2E
 
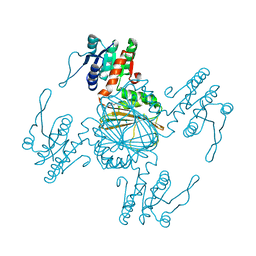 | |
4IHL
 
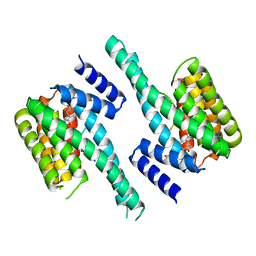 | | Human 14-3-3 isoform zeta in complex with a diphoyphorylated C-RAF peptide and Cotylenin A | | Descriptor: | (1R,3aS,4R,5R,6R,9aR,10E)-6-({(1S,2R,4S,5R,6R,8S,9S)-5-hydroxy-2-(methoxymethyl)-9-methyl-9-[(2S)-oxiran-2-yl]-3,7,10,1 1-tetraoxatricyclo[6.2.1.0~1,6~]undec-4-yl}oxy)-1-(methoxymethyl)-4,9a-dimethyl-7-(propan-2-yl)-1,2,3,3a,4,5,6,8,9,9a-de cahydrodicyclopenta[a,d][8]annulene-1,5-diol, 14-3-3 protein zeta/delta, POTASSIUM ION, ... | | Authors: | Molzan, M, Ottmann, C. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of Physical RAF/14-3-3 Interaction by Cotylenin A as Treatment Strategy for RAS Mutant Cancers.
Acs Chem.Biol., 8, 2013
|
|
3UBW
 
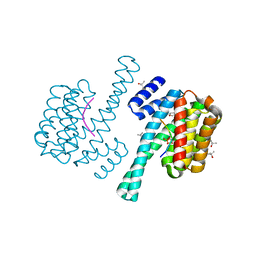 | | Complex of 14-3-3 isoform epsilon, a Mlf1 phosphopeptide and a small fragment hit from a FBDD screen | | Descriptor: | (3S)-pyrrolidin-3-ol, 14-3-3 protein epsilon, Myeloid leukemia factor 1, ... | | Authors: | Molzan, M, Weyand, M, Rose, R, Ottmann, C. | | Deposit date: | 2011-10-25 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the MLF1/14-3-3 interaction.
Febs J., 279, 2012
|
|
1RVF
 
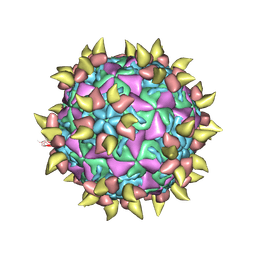 | | FAB COMPLEXED WITH INTACT HUMAN RHINOVIRUS | | Descriptor: | FAB 17-IA, HUMAN RHINOVIRUS 14 COAT PROTEIN | | Authors: | Smith, T.J. | | Deposit date: | 1996-09-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon.
Nature, 383, 1996
|
|
8VX0
 
 | | CRYSTAL STRUCTURE OF CYP2C9*14 IN COMPLEX WITH LOSARTAN | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Shah, M.B. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and biophysical analysis of cytochrome P450 2C9*14 and *27 variants in complex with losartan.
J.Inorg.Biochem., 258, 2024
|
|
3IQJ
 
 | | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (10mer) | | Descriptor: | 10-mer peptide from RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Ottmann, C, Weyand, M. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol.Cell.Biol., 30, 2010
|
|
8QTT
 
 | |
8QTF
 
 | |
6FBY
 
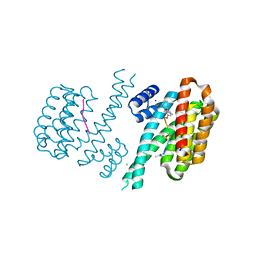 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2b with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methylphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
5JLC
 
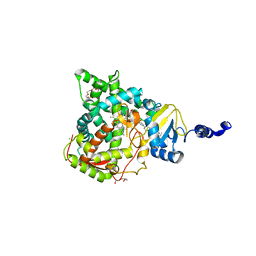 | | Structure of CYP51 from the pathogen Candida glabrata | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, CHLORIDE ION, Lanosterol 14-alpha demethylase, ... | | Authors: | Keniya, M.V, Sabherwal, M, Wilson, R.K, Sagatova, A.A, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2016-04-26 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Full-Length Lanosterol 14 alpha-Demethylases of Prominent Fungal Pathogens Candida albicans and Candida glabrata Provide Tools for Antifungal Discovery.
Antimicrob.Agents Chemother., 62, 2018
|
|
3TSG
 
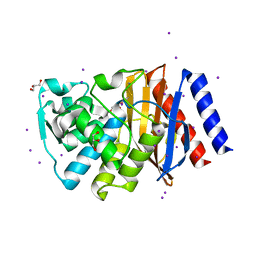 | | Crystal structure of GES-14 | | Descriptor: | Extended-spectrum beta-lactamase GES-14, GLYCEROL, IODIDE ION | | Authors: | Delbruck, H, Hoffmann, K.M.V, Bebrone, C. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and crystallographic studies of extended-spectrum GES-11, GES-12, and GES-14 beta-lactamases.
Antimicrob.Agents Chemother., 56, 2012
|
|
3EFZ
 
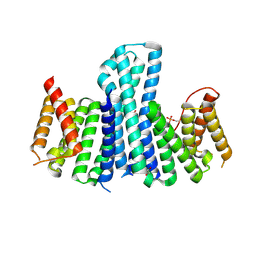 | | Crystal Structure of a 14-3-3 protein from cryptosporidium parvum (cgd1_2980) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein | | Authors: | Wernimont, A.K, Dong, A, Qiu, W, Lew, J, Wasney, G.A, Vedadi, M, Kozieradzki, I, Zhao, Y, Ren, H, Alam, Z, Lin, Y.H, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
6FAU
 
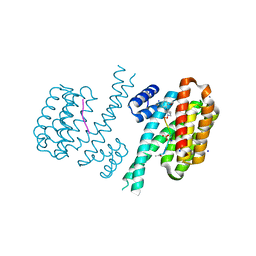 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2e-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(2-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FI4
 
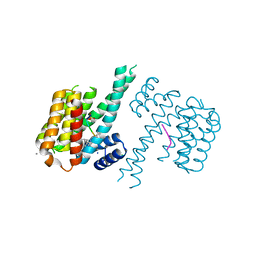 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2e with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FAV
 
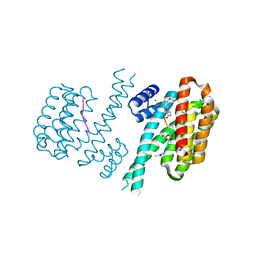 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FI5
 
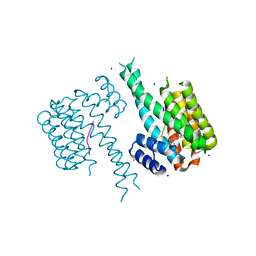 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2d with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FAW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2c-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-[2-(2-methoxyethoxy)phenyl]-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
7NUM
 
 | | Rhinovirus-14 ICAM-1 empty particle at pH 6.2 | | Descriptor: | Genome polyprotein, P1 | | Authors: | Hrebik, D, Plevka, P. | | Deposit date: | 2021-03-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NUO
 
 | | Rhinovirus 14 empty particle at pH 6.2 | | Descriptor: | Genome polyprotein, P1 | | Authors: | Hrebik, D, Plevka, P. | | Deposit date: | 2021-03-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NUN
 
 | | Rhinovirus 14 ICAM-1 virion-like particle at pH 6.2 | | Descriptor: | Genome polyprotein, Octanucleotide | | Authors: | Hrebik, D, Plevka, P. | | Deposit date: | 2021-03-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
