7JXC
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | NONAETHYLENE GLYCOL, S2H14 antigen-binding (Fab) fragment | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
1UHB
 
 | | Crystal structure of porcine alpha trypsin bound with auto catalyticaly produced native peptide at 2.15 A resolution | | Descriptor: | 9-mer peptide from Trypsin, ACETATE ION, CALCIUM ION, ... | | Authors: | Pattabhi, V, Syed Ibrahim, B, Shamaladevi, N. | | Deposit date: | 2003-06-27 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Trypsin activity reduced by an autocatalytically produced nonapeptide.
J.Biomol.Struct.Dyn., 21, 2004
|
|
7ICE
 
 | |
7ICO
 
 | |
7K12
 
 | | ACMSD in complex with diflunisal | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, CITRIC ACID, ... | | Authors: | Yang, Y, Liu, A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Diflunisal Derivatives as Modulators of ACMS Decarboxylase Targeting the Tryptophan-Kynurenine Pathway.
J.Med.Chem., 64, 2021
|
|
1CH8
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH A STRINGENT EFFECTOR, PPG2':3'P | | Descriptor: | GUANOSINE 5'-DIPHOSPHATE 2':3'-CYCLIC MONOPHOSPHATE, HADACIDIN, INOSINIC ACID, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
7JK6
 
 | | Structure of Drosophila ORC in the active conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Schmidt, J.M, Bleichert, F. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
1CKW
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
7JNY
 
 | | Crystal structure of CXCL13 | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Rosenberg Jr, E.M, Rajasekaran, D, Murphy, J.W, Pantouris, G, Lolis, E.J. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N-terminal length and side-chain composition of CXCL13 affect crystallization, structure and functional activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7JO1
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butyramide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butanamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
1ODT
 
 | | cephalosporin C deacetylase mutated, in complex with acetate | | Descriptor: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
1OE9
 
 | | Crystal structure of Myosin V motor with essential light chain-nucleotide-free | | Descriptor: | MYOSIN LIGHT CHAIN 1, SLOW-TWITCH MUSCLE A ISOFORM, MYOSIN VA, ... | | Authors: | Coureux, P.-D, Wells, A.L, Menetrey, J, Yengo, C.M, Morris, C.A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2003-03-21 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural State of the Myosin V Motor without Bound Nucleotide
Nature, 425, 2003
|
|
1OC4
 
 | | Lactate dehydrogenase from Plasmodium berghei | | Descriptor: | GLYCEROL, L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Winter, V.J, Brady, R.L. | | Deposit date: | 2003-02-05 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Plasmodium Berghei Lactate Dehydrogenase Indicates the Unique Structural Differences of These Enzymes are Shared Across the Plasmodium Genus
Mol.Biochem.Parasitol., 131, 2003
|
|
1CKZ
 
 | | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR: SOLUTION STRUCTURES OF PEPTIDES BASED ON THE PHE508 REGION, THE MOST COMMON SITE OF DISEASE-CAUSING DELTA-F508 MUTATION | | Descriptor: | PROTEIN (CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR (CFTR)) | | Authors: | Massiah, M.A, Ko, Y.H, Pedersen, P.L, Mildvan, A.S. | | Deposit date: | 1999-04-26 | | Release date: | 1999-05-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation.
Biochemistry, 38, 1999
|
|
1O7L
 
 | |
1O6E
 
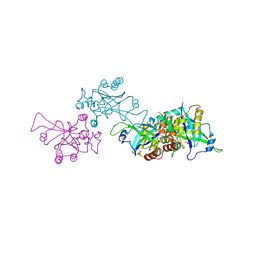 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
1CYY
 
 | |
1S84
 
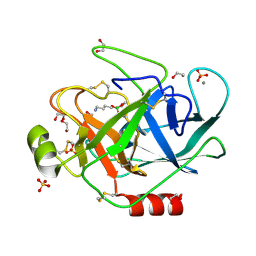 | | PORCINE TRYPSIN COVALENT COMPLEX WITH 4-AMINO BUTANOL, BORATE AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3,2-DIOXABOROLAN-2-YLOXY)BUTAN-1-AMINIUM, CALCIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1OD7
 
 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-(naphthyl-2-carbonyl)-amino-9-deoxy-Neu5Ac (NAP compound) | | Descriptor: | ME-A-9-N-(NAPHTHYL-2-CARBONYL)-AMINO-9-DEOXY-NEU5AC, SIALOADHESIN | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1SGE
 
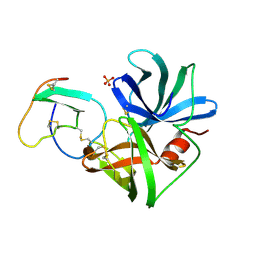 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1SGQ
 
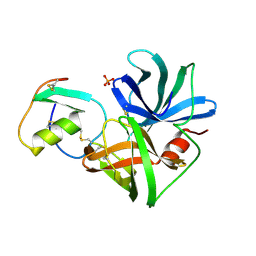 | |
2ANB
 
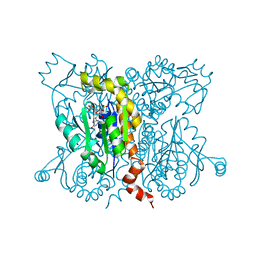 | | Crystal Structure Of Oligomeric E.coli Guanylate Kinase In Complex With GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, SULFATE ION | | Authors: | Hible, G, Renault, L, Schaeffer, F, Christova, P, Radulescu, A.Z, Evrin, C, Gilles, A.M, Cherfils, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase
J.Mol.Biol., 352, 2005
|
|
1NPM
 
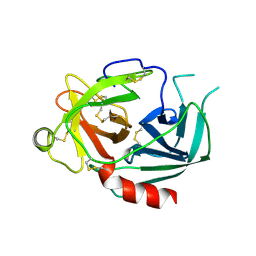 | | NEUROPSIN, A SERINE PROTEASE EXPRESSED IN THE LIMBIC SYSTEM OF MOUSE BRAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUROPSIN | | Authors: | Kishi, T, Kato, M, Shimizu, T, Kato, K, Matsumoto, K, Yoshida, S, Shiosaka, S, Hakoshima, T. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis.
J.Biol.Chem., 274, 1999
|
|
1NPW
 
 | | Crystal structure of HIV protease complexed with LGZ479 | | Descriptor: | CARBAMIC ACID 1-{5-BENZYL-5-[2-HYDROXY-4-PHENYL-3-(TETRAHYDRO-FURAN- 3-YLOXYCARBONYLAMINO)-BUTYL]-4-OXO-4,5-DIHYDRO-1H-PYRROL-3-YL}- INDAN-2-YL ESTER, POL polyprotein | | Authors: | Smith III, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
1SC9
 
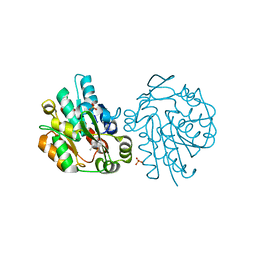 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | Descriptor: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
