1XLR
 
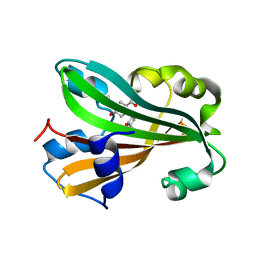 | | CHORISMATE LYASE WITH INHIBITOR VANILLATE | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Chorismate--pyruvate lyase | | Authors: | Gallagher, D.T, Smith, N. | | Deposit date: | 2004-09-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
1PQI
 
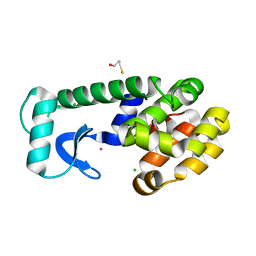 | | T4 LYSOZYME CORE REPACKING MUTANT I118L/CORE7/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1C90
 
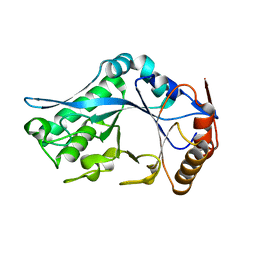 | | Endo-Beta-N-Acetylglucosaminidase H, E132Q Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Assp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1PV1
 
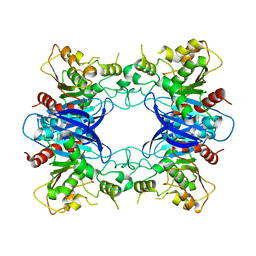 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | Descriptor: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | Authors: | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-26 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
1PNZ
 
 | |
1RBA
 
 | |
1PQO
 
 | | T4 Lysozyme Core Repacking Mutant L118I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1YF2
 
 | | Three-dimensional structure of DNA sequence specificity (S) subunit of a type I restriction-modification enzyme and its functional implications | | Descriptor: | Type I restriction-modification enzyme, S subunit | | Authors: | Kim, J.S, Degiovanni, A, Jancarik, J, Adams, P.D, Yokota, H.A, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-12-30 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1O35
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-4-FLUOROBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
2BRI
 
 | |
1O3C
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O3K
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-BROMO-4-METHYLBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
2C4B
 
 | | Inhibitor cystine knot protein McoEeTI fused to the catalytically inactive barnase mutant H102A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BARNASE MCOEETI FUSION, ... | | Authors: | Niemann, H.H, Schmoldt, H.U, Wentzel, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2005-10-18 | | Release date: | 2005-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Barnase Fusion as a Tool to Determine the Crystal Structure of the Small Disulfide-Rich Protein Mcoeeti.
J.Mol.Biol., 356, 2006
|
|
1C92
 
 | | Endo-Beta-N-Acetylglucosaminidase H, E132A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1RGY
 
 | | Citrobacter freundii GN346 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
1AET
 
 | | VARIATION IN THE STRENGTH OF A CH TO O HYDROGEN BOND IN AN ARTIFICIAL PROTEIN CAVITY (1-METHYLIMIDAZOLE) | | Descriptor: | 1-METHYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1Y6W
 
 | | Trapped intermediate of calmodulin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin, ... | | Authors: | Grabarek, Z. | | Deposit date: | 2004-12-07 | | Release date: | 2005-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Trapped Intermediate of Calmodulin: Calcium Regulation of EF-hand Proteins from a New Perspective.
J.Mol.Biol., 346, 2005
|
|
180D
 
 | |
1Y92
 
 | | Crystal structure of the P19A/N67D Variant Of Bovine seminal Ribonuclease | | Descriptor: | Seminal ribonuclease | | Authors: | Picone, D, Di Fiore, A, Ercole, C, Franzese, M, Sica, F, Tomaselli, S, Mazzarella, L. | | Deposit date: | 2004-12-14 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of the Hinge Loop in Domain Swapping: THE SPECIAL CASE OF BOVINE SEMINAL RIBONUCLEASE.
J.Biol.Chem., 280, 2005
|
|
1RRK
 
 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1NN8
 
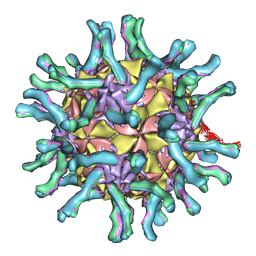 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
1RS0
 
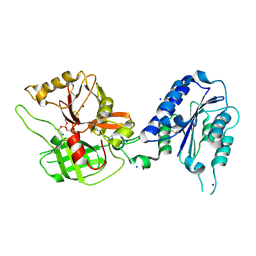 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with Di-isopropyl-phosphate (DIP) | | Descriptor: | Complement factor B, DIISOPROPYL PHOSPHONATE, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
181D
 
 | |
1NU9
 
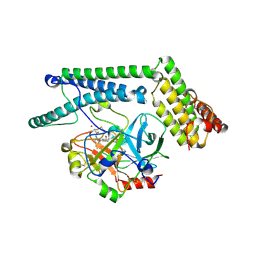 | | Staphylocoagulase-Prethrombin-2 complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
2AZS
 
 | | NMR structure of the N-terminal SH3 domain of Drk (calculated without NOE restraints) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
