8ED7
 
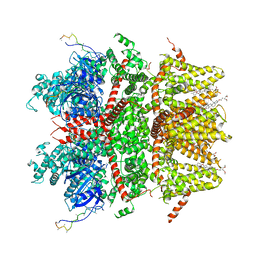 | | cryo-EM structure of TRPM3 ion channel in apo state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Transient receptor potential cation channel, subfamily M, ... | | Authors: | Zhao, C, MacKinnon, R. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional analyses of a GPCR-inhibited ion channel TRPM3.
Neuron, 111, 2023
|
|
8ED6
 
 | |
8ED5
 
 | |
8ED4
 
 | | Structure of the complex between the arsenite oxidase and its native electron acceptor cytochrome c552 from Pseudorhizobium sp. str. NT-26 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, AroA, AroB, ... | | Authors: | Maher, M.J, Poddar, N. | | Deposit date: | 2022-09-03 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the complex between the arsenite oxidase from Pseudorhizobium banfieldiae sp. strain NT-26 and its native electron acceptor cytochrome c 552.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECX
 
 | |
8ECW
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 11 | | Descriptor: | (2R,3R,4R,5S)-1-{2-[4-(2-{[(5M)-3-chloro-5-(pyridazin-3-yl)phenyl]amino}ethyl)phenyl]ethyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ECR
 
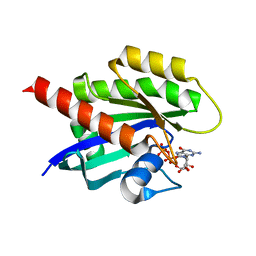 | | KRAS4b WT 1-185 bound to GDP-Mg2+ | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Betts, L, Rossman, K.L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | KRAS4b WT 1-185 bound to GDP-Mg2+
To Be Published
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECP
 
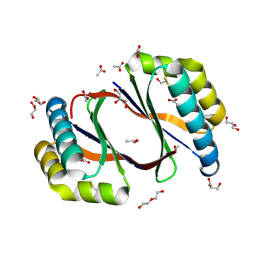 | |
8ECO
 
 | | Microbacterium phage Oxtober96 | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECN
 
 | | Mycobacterium phage Ogopogo | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECK
 
 | | Gordonia phage Cozz | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECJ
 
 | | Mycobacterium phage Cain | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECI
 
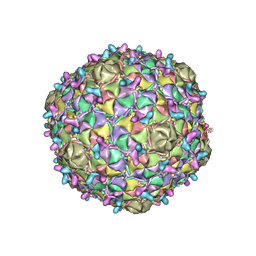 | | Arthrobacter phage Bridgette | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECG
 
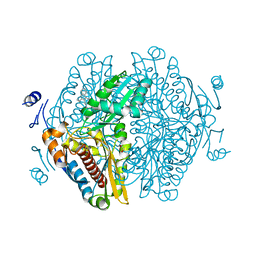 | | Complex of HMG1 with pitavastatin | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | Authors: | Haywood, J, Bond, C.S. | | Deposit date: | 2022-09-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
8ECF
 
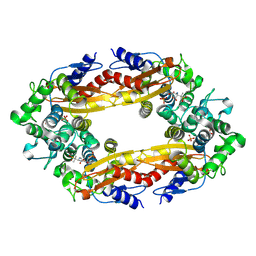 | |
8ECD
 
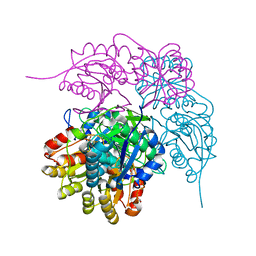 | | E. coli L-asparaginase II mutant (V27T) in complex with L-Asp | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, CITRIC ACID, ... | | Authors: | Strzelczyk, P, Wlodawer, A, Lubkowski, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.
Febs Lett., 596, 2022
|
|
8EC8
 
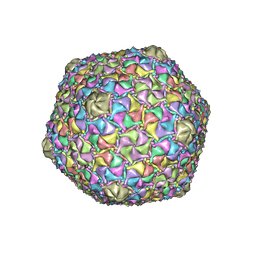 | | Mycobacterium phage Bobi | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC6
 
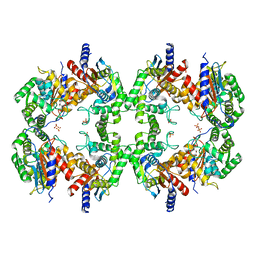 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EC4
 
 | |
8EC2
 
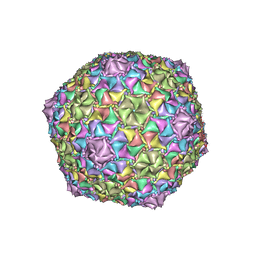 | | Mycobacterium phage Adephagia | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC1
 
 | |
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | Descriptor: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBX
 
 | |
