3O5T
 
 | | Structure of DraG-GlnZ complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dinitrogenase reductase activacting glicohydrolase, MAGNESIUM ION, ... | | Authors: | Rajendran, C, Li, X.-D, Winkler, F.K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of the GlnZ-DraG complex reveals a different form of PII-target interaction
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O7O
 
 | |
4PXM
 
 | |
3D9I
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, GLYCEROL, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9O
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, CTD-PEPTIDE, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9L
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | ACETATE ION, CTD-PEPTIDE, GLYCEROL, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9M
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, CTD-PEPTIDE, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
6A8I
 
 | | Crystal structure of endo-arabinanase ABN-TS D147N mutant in complex with arabinohexaose | | Descriptor: | CALCIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, endo-alpha-(1->5)-L-arabinanase | | Authors: | Yamaguchi, A, Tada, T. | | Deposit date: | 2018-07-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of endo-1,5-alpha-L-arabinanase mutants from Bacillus thermodenitrificans TS-3 in complex with arabino-oligosaccharides.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5EXE
 
 | | Crystal structure of oxalate oxidoreductase from Moorella thermoacetica bound with carboxy-TPP adduct | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | One-carbon chemistry of oxalate oxidoreductase captured by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7AA2
 
 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with ABTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3TSO
 
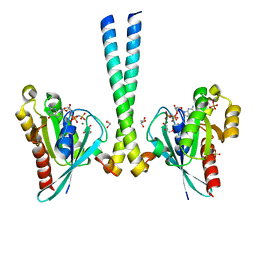 | | Structure of the cancer associated Rab25 protein in complex with FIP2 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Oda, S, Lall, P.Y, McCaffrey, M.W, Khan, A.R. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the cancer associated Rab25 protein in complex with FIP2
TO BE PUBLISHED
|
|
6A8H
 
 | | Crystal structure of endo-arabinanase ABN-TS D27A mutant in complex with arabinotriose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, ... | | Authors: | Yamaguchi, A, Tada, T. | | Deposit date: | 2018-07-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of endo-1,5-alpha-L-arabinanase mutants from Bacillus thermodenitrificans TS-3 in complex with arabino-oligosaccharides.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7VSI
 
 | | Structure of human SGLT2-MAP17 complex bound with empagliflozin | | Descriptor: | (2S,3R,4R,5S,6R)-2-[4-chloranyl-3-[[4-[(3S)-oxolan-3-yl]oxyphenyl]methyl]phenyl]-6-(hydroxymethyl)oxane-3,4,5-triol, PALMITIC ACID, PDZK1-interacting protein 1, ... | | Authors: | Chen, L, Niu, Y, Liu, R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-12-15 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter.
Nature, 601, 2022
|
|
3UI4
 
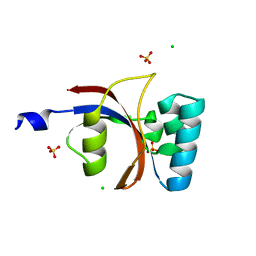 | | 0.8 A resolution crystal structure of human Parvulin 14 | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 4, SULFATE ION | | Authors: | Mueller, J.W, Link, N.M, Matena, A, Hoppstock, L, Rueppel, A, Bayer, P, Blankenfeldt, W. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases.
J.Am.Chem.Soc., 133, 2011
|
|
3VHS
 
 | |
2RUD
 
 | |
4R8P
 
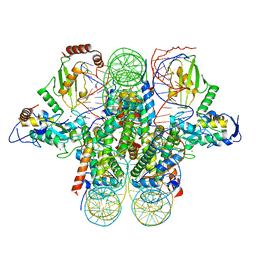 | | Crystal structure of the Ring1B/Bmi1/UbcH5c PRC1 ubiquitylation module bound to the nucleosome core particle | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase RING2, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | McGinty, R.K, Henrici, R.C, Tan, S. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2846 Å) | | Cite: | Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome.
Nature, 514, 2014
|
|
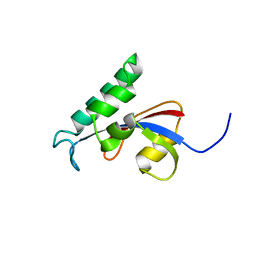
2RUC
 
 | |
3IKD
 
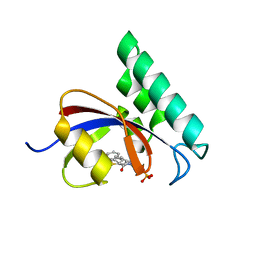 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
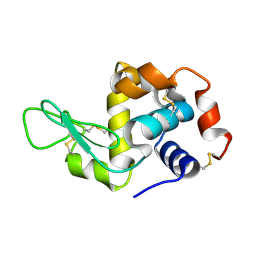
8H3W
 
 | |
3MMW
 
 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
3MMU
 
 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase, NICKEL (II) ION | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
4JXM
 
 | | Crystal structure of RRP9 WD40 repeats | | Descriptor: | U3 small nucleolar RNA-interacting protein 2, UNKNOWN ATOM OR ION | | Authors: | Wu, X, Tempel, W, Xu, C, El Bakkouri, M, He, H, Seitova, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of RRP9 WD40 repeats
To be Published
|
|
3IK8
 
 | |
3IKG
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | Descriptor: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-(3-methylphenyl)propyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Parge, H, Ferre, R.A, Greasley, S, Matthews, D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
