5EOW
 
 | | Crystal Structure of 6-Hydroxynicotinic Acid 3-Monooxygenase from Pseudomonas putida KT2440 | | Descriptor: | 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yuen, M.E, Zhen, W, Gerwig, T.J, Story, R.W, Kopp, M, Nakamoto, K, Snider, M.J, Hicks, K.A. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-08 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Characterization of 6-Hydroxynicotinic Acid 3-Monooxygenase, A Novel Decarboxylative Hydroxylase Involved in Aerobic Nicotinate Degradation.
Biochemistry, 55, 2016
|
|
5EVY
 
 | | Salicylate hydroxylase substrate complex | | Descriptor: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | Authors: | Morimoto, Y, Uemura, T. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
3RP7
 
 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD and uric acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIC ACID, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
3RP6
 
 | | Crystal Structure of Klebsiella pneumoniae HpxO complexed with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, flavoprotein monooxygenase | | Authors: | Hicks, K.A, O'Leary, S.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-16 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Studies of HpxO, a Novel Flavin Adenine Dinucleotide-Dependent Urate Oxidase from Klebsiella pneumoniae.
Biochemistry, 52, 2013
|
|
5X6Q
 
 | | Crystal structure of Pseudomonas fluorescens KMO in complex with Ro 61-8048 | | Descriptor: | 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6R
 
 | |
8WVB
 
 | | Crystal structure of Lsd18 mutant S195M | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
8WVF
 
 | | Crystal structure of Lsd18 mutant T189M and S195M | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.757 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
5X68
 
 | | Crystal Structure of Human KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
5X6P
 
 | | Crystal structure of Pseudomonas fluorescens KMO | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Inhibitor-Induced Hydrogen Peroxide Production by Kynurenine 3-Monooxygenase
Cell Chem Biol, 25, 2018
|
|
3GMC
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase with substrate bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-6-methylpyridine-3-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
3GMB
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
6DLL
 
 | | 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of p-hydroxybenzoate hydroxylase (PobA) from Pseudomonas putida with PobA from other Pseudomonas spp. and other monooxygenases.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
8Y2S
 
 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | Descriptor: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2024-01-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
6BZ5
 
 | |
6LKD
 
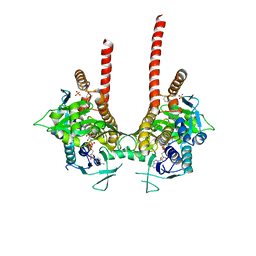 | | in meso full-length rat KMO in complex with a pyrazoyl benzoic acid inhibitor | | Descriptor: | 5-[5-(4-chloranyl-3-fluoranyl-phenyl)-4-methyl-pyrazol-1-yl]-2-phenylmethoxy-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
6LKE
 
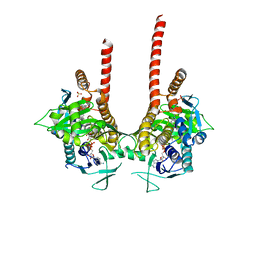 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
6JU1
 
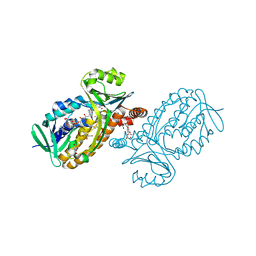 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
2BRA
 
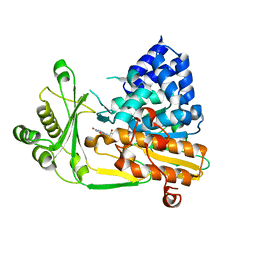 | | Structure of N-Terminal FAD Binding motif of mouse MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Nadella, M, Bianchet, M.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the axon guidance protein MICAL.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
6SW1
 
 | | Crystal Structure of P. aeruginosa PqsL: R41Y, I43R, G45R, C105G mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mattevi, A, Rovida, S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoinduced monooxygenation involving NAD(P)H-FAD sequential single-electron transfer.
Nat Commun, 11, 2020
|
|
6N04
 
 | | The X-ray crystal structure of AbsH3, an FAD dependent reductase from the Abyssomicin biosynthesis pathway in Streptomyces | | Descriptor: | AbsH3, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Clinger, J.A, Wang, X, Cai, W, Miller, M.D, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-11-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The crystal structure of AbsH3: A putative flavin adenine dinucleotide-dependent reductase in the abyssomicin biosynthesis pathway.
Proteins, 2020
|
|
6PVF
 
 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVH
 
 | | Crystal structure of PhqK in complex with paraherquamide K | | Descriptor: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVG
 
 | | Crystal structure of ligand free PhqK | | Descriptor: | FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
