5JVR
 
 | |
9D0V
 
 | | Crystal structure of CDK2/CyclinE1 in complex with Cpd 2 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-8-cyclopentyl-2-[4-(ethanesulfonyl)-2-methylanilino]pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, ... | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
1KZ0
 
 | |
7CKR
 
 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate BAY-8002 in the outward-open conformation. | | Descriptor: | 2-[[2-chloranyl-5-(phenylsulfonyl)phenyl]carbonylamino]benzoic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6ZM2
 
 | |
8Q7Z
 
 | | Structure of the G. gallus 80S non-rotated ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Nurullina, L, Jenner, L, Yusupov, M. | | Deposit date: | 2023-08-17 | | Release date: | 2024-08-28 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the inactive ribosome complex accumulated in chick embryo cells in cold-stress conditions.
Febs Lett., 598, 2024
|
|
6BX6
 
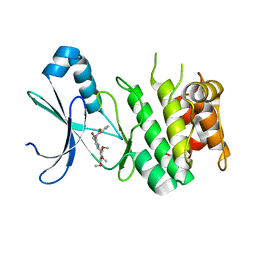 | | AMP-Activated protein kinase (AMPK) inhibition by SBI-0206965: alpha 2 kinase domain bound to SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Dite, T.A, Langendorf, C.G, Scott, J.W, Oakhill, J.S. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965.
J. Biol. Chem., 293, 2018
|
|
4RSC
 
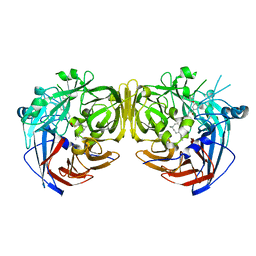 | | Crystal structure of RPE65 in complex with emixustat and palmitate | | Descriptor: | (1R)-3-amino-1-[3-(cyclohexylmethoxy)phenyl]propan-1-ol, FE (II) ION, PALMITIC ACID, ... | | Authors: | Kiser, P.D, Shi, W, Palczewski, K. | | Deposit date: | 2014-11-07 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic mechanism of a retinoid isomerase essential for vertebrate vision.
Nat.Chem.Biol., 11, 2015
|
|
6ILD
 
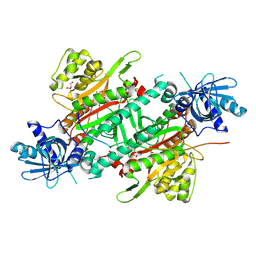 | | Crystal Structure of Human LysRS: P38/AIMP2 Complex II | | Descriptor: | 5'-O-[(S)-hydroxy(methyl)phosphoryl]adenosine, Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, GLYCEROL, ... | | Authors: | Hei, Z, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2018-10-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex.
J. Biol. Chem., 294, 2019
|
|
4RX1
 
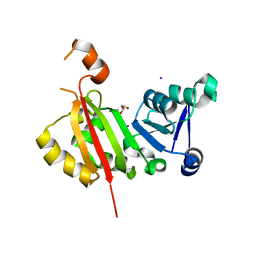 | | Crystal Structure of antibiotic-resistance methyltransferase Kmr | | Descriptor: | GLYCEROL, IODIDE ION, Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
6INK
 
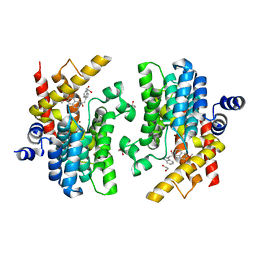 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
5X27
 
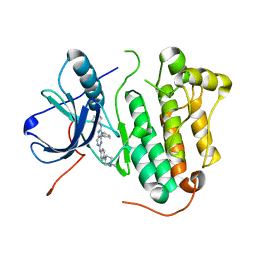 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
9FVP
 
 | |
8OF1
 
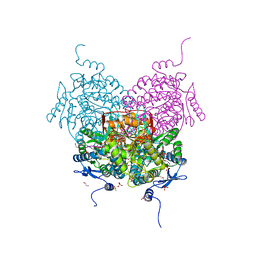 | | Structure of ALDH5F1 from moss Physcomitrium patens in complex with NAD+ in the contracted conformation | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
6RCY
 
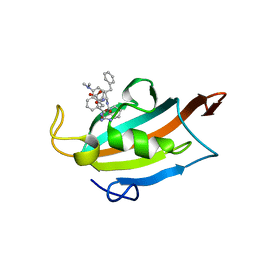 | |
7AIA
 
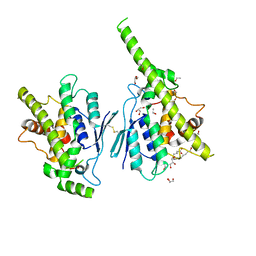 | | Complex of human GDAP1 with hexadecanedioic acid | | Descriptor: | ETHANOL, GLYCEROL, Ganglioside-induced differentiation-associated protein 1, ... | | Authors: | Nguyen, G.T.T, Sutinen, A, Kursula, P. | | Deposit date: | 2020-09-26 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Complete Dimeric Human GDAP1 Core Domain Provides Insights into Ligand Binding and Clustering of Disease Mutations.
Front Mol Biosci, 7, 2020
|
|
8W89
 
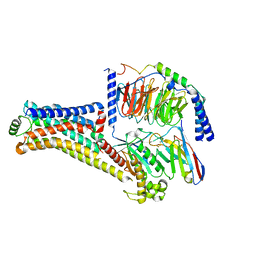 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6RDC
 
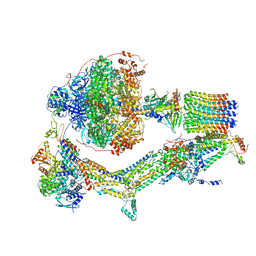 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 2, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDY
 
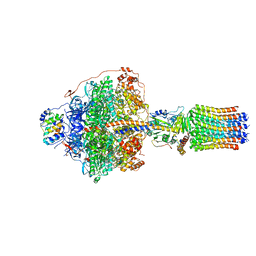 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1F, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
2UYJ
 
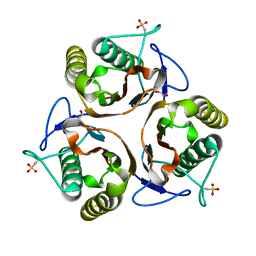 | | Crystal structure of E. coli TdcF with bound ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
5N58
 
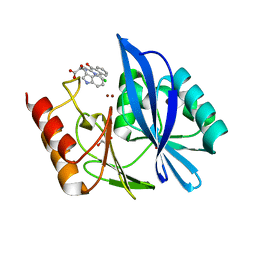 | | di-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, GLYCEROL, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
9G7B
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and loreclezole | | Descriptor: | 1-[(~{Z})-2-chloranyl-2-(2,4-dichlorophenyl)ethenyl]-1,2,4-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-07-20 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | CryoEM structure of human rho1 GABAA receptor in complex with neurosteroid
To be published
|
|
6RED
 
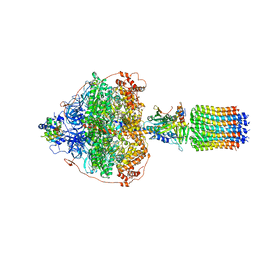 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
8OKJ
 
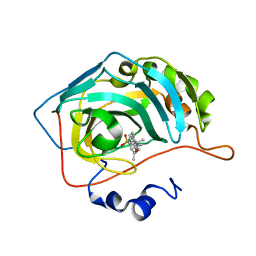 | | Carbonic Anhydrase IX like mutant in Complex with Steriod_Sulphamoyl inhibitor AKI_12 | | Descriptor: | Carbonic anhydrase 2, ZINC ION, [(3~{R},5~{S},8~{R},9~{S},10~{S},13~{S},14~{S},17~{S})-17-ethanoyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl] sulfamate | | Authors: | Brynda, J, Rezacova, P.M, Kudova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Carbonic Anhydrase II in Complex with Steriod_Sulphamoyl
To Be Published
|
|
6RDJ
 
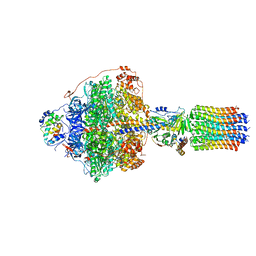 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
