3UEI
 
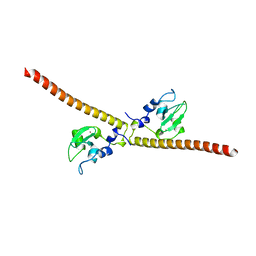 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
5SZC
 
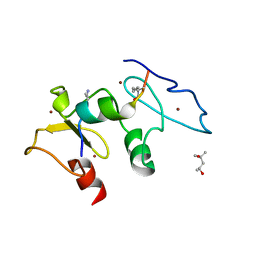 | | Structure of human Dpf3 double-PHD domain bound to histone H3 tail peptide with monomethylated K4 and acetylated K14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histone H3 tail peptide, ZINC ION, ... | | Authors: | Singh, N, Local, A, Shiau, A, Ren, B, Corbett, K.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Identification of H3K4me1-associated proteins at mammalian enhancers.
Nat. Genet., 50, 2018
|
|
2C5Z
 
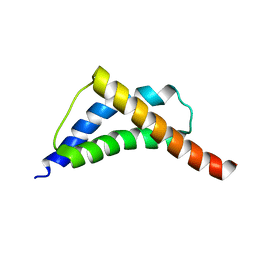 | | Structure and CTD binding of the Set2 SRI domain | | Descriptor: | SET DOMAIN PROTEIN 2 | | Authors: | Vojnic, E, Simon, B, Strahl, B.D, Sattler, M, Cramer, P. | | Deposit date: | 2005-11-03 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription.
J. Biol. Chem., 281, 2006
|
|
5YJE
 
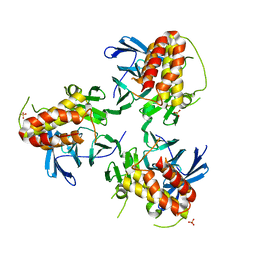 | | Crystal structure of HIRA(644-1017) | | Descriptor: | Protein HIRA, SULFATE ION | | Authors: | Sato, Y, Senda, M, Senda, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit
Nat Commun, 9, 2018
|
|
3IJY
 
 | | Structure of S67-27 in Complex with Kdo(2.8)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3IJH
 
 | | Structure of S67-27 in Complex with Ko | | Descriptor: | Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), MAGNESIUM ION, ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3IJS
 
 | | Structure of S67-27 in Complex with TSBP | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-alpha-D-glucopyranose, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
3IKC
 
 | | Structure of S67-27 in Complex with Kdo(2.8)-7-O-methyl-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-(1E)-prop-1-en-1-yl 3-deoxy-7-O-methyl-alpha-D-manno-oct-2-ulopyranosidonic acid, Immunoglobulin heavy chain (IGG3), Immunoglobulin light chain (IGG3), ... | | Authors: | Brooks, C.L, Blackler, R.J, Evans, S.V. | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen.
Glycobiology, 20, 2010
|
|
2F5K
 
 | |
8G2K
 
 | |
3KMT
 
 | | Crystal structure of vSET/SAH/H3 ternary complex | | Descriptor: | A612L protein, Histone H3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wei, H, Zhou, M.M. | | Deposit date: | 2009-11-11 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dimerization of a viral SET protein endows its function.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MP1
 
 | | Complex structure of Sgf29 and trimethylated H3K4 | | Descriptor: | ACETATE ION, H3K4me3 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, ... | | Authors: | Li, J, Ruan, J, Wu, M, Xue, X, Zang, J. | | Deposit date: | 2010-04-24 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
2RSD
 
 | | Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice | | Descriptor: | E3 SUMO-protein ligase SIZ1, ZINC ION | | Authors: | Shindo, H, Tsuchiya, W, Suzuki, R, Yamazaki, T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2
Febs Lett., 586, 2012
|
|
2KTM
 
 | | Solution NMR structure of H2H3 domain of ovine prion protein (residues 167-234) | | Descriptor: | Major prion protein | | Authors: | Pastore, A, Adrover, M, Pauwels, K, de Chiara, C, Prigent, S, Rezeai, H. | | Deposit date: | 2010-02-04 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Prion fibrillization is mediated by a native structural element that comprises helices H2 and H3.
J.Biol.Chem., 285, 2010
|
|
1QST
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
1QSR
 
 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND ACETYL-COENZYME A | | Descriptor: | ACETYL COENZYME *A, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
2YYR
 
 | |
3QLC
 
 | |
8TX3
 
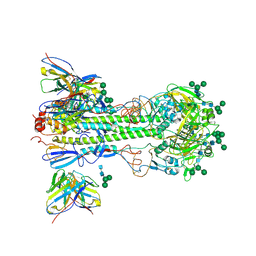 | | Fab 3864-6 in complex with influenza HA H3-VIC11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 3864-6 Heavy Chain, Fab 3864-6 Light Chain, ... | | Authors: | Morano, N.C, Shapiro, L. | | Deposit date: | 2023-08-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Fab 3864-6 in complex with influenza HA H3-VIC11
To Be Published
|
|
5HJB
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K9 | | Descriptor: | Protein AF-9, peptide of Histone H3.1 | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-12 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
5HJD
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K18 | | Descriptor: | COPPER (II) ION, Protein AF-9, SULFATE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain.
Mol.Cell, 62, 2016
|
|
5HJC
 
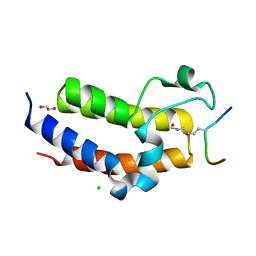 | | BRD3 second bromodomain in complex with histone H3 acetylation at K18 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, CHLORIDE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
8JWS
 
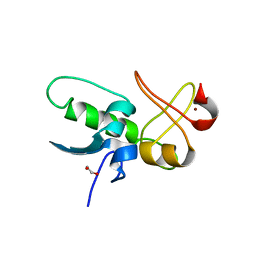 | | ePHD domain of PHD Finger Protein 7 (PHF7) | | Descriptor: | 1,2-ETHANEDIOL, PHD finger protein 7, ZINC ION | | Authors: | Bang, I, Lee, H.S, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
5SZB
 
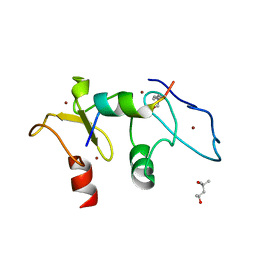 | | Structure of human Dpf3 double-PHD domain bound to histone H3 tail peptide with acetylated K14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histone H3 tail peptide, ZINC ION, ... | | Authors: | Singh, N, Local, A, Shiau, A, Ren, B, Corbett, K.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification of H3K4me1-associated proteins at mammalian enhancers.
Nat. Genet., 50, 2018
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
