1N3X
 
 | | Ligand-free High-Affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2002-10-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
2H25
 
 | |
1PUE
 
 | | PU.1 ETS DOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*GP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*CP*CP*TP*TP*TP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR PU.1 (TF PU.1)) | | Authors: | Kodandapani, R, Pio, F, Ni, C.Z, Piccialli, G, Klemsz, M, McKercher, S, Maki, R.A, Ely, K.R. | | Deposit date: | 1996-07-08 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Nature, 380, 1996
|
|
4RWF
 
 | |
2H8O
 
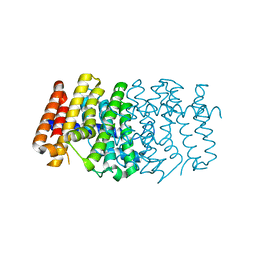 | | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens | | Descriptor: | Geranyltranstransferase | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2H8B
 
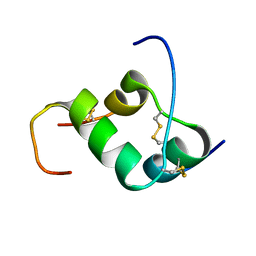 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
2K6T
 
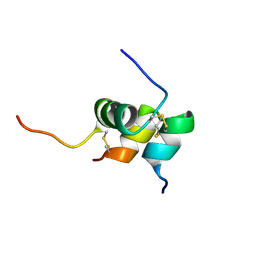 | | Solution structure of the relaxin-like factor | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
1RTR
 
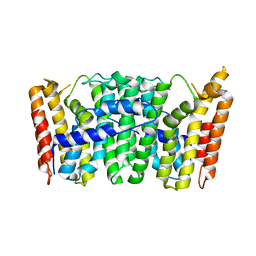 | | Crystal Structure of S. Aureus Farnesyl Pyrophosphate Synthase | | Descriptor: | geranyltranstransferase | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Mol.Biol., 279, 2004
|
|
4PQK
 
 | | C-Terminal domain of DNA binding protein | | Descriptor: | Maltose ABC transporter periplasmic protein, Truncated replication protein RepA, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N.K. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1N3W
 
 | | Engineered High-Affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2002-10-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
1N4C
 
 | | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin | | Descriptor: | Auxilin | | Authors: | Gruschus, J.M, Han, C.J, Greener, T, Greene, L.E, Ferretti, J.A, Eisenberg, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles.
Biochemistry, 43, 2004
|
|
2KLF
 
 | | PERE NMR structure of maltodextrin-binding protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2KLX
 
 | |
1OMP
 
 | |
4PE2
 
 | | MBP PilA1 CD160 | | Descriptor: | MALONATE ION, Maltose ABC transporter periplasmic protein,Prepilin-type N-terminal cleavage/methylation domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-04-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
4TSM
 
 | | MBP-fusion protein of PilA1 from C. difficile R20291 residues 26-166 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein, pilin chimera, ... | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-06-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
1T95
 
 | |
4RWG
 
 | |
1T0K
 
 | | Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex | | Descriptor: | 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3', 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3', 60S ribosomal protein L30, ... | | Authors: | Chao, J.A, Williamson, J.R. | | Deposit date: | 2004-04-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Structure, 12, 2004
|
|
6WH0
 
 | | Crystal structure of HyBcl-2-4 with HyBax BH3 | | Descriptor: | Apoptosis regulator BAX, Maltodextrin-binding protein,Bcl-2-like 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kvansakul, M, Hinds, M.G, Banjara, S. | | Deposit date: | 2020-04-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structural basis of Bcl-2 mediated cell death regulation in hydra.
Biochem.J., 477, 2020
|
|
6WGZ
 
 | |
6XDS
 
 | |
6XRX
 
 | | Crystal structure of the mosquito protein AZ1 as an MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Foo, A.C.Y. | | Deposit date: | 2020-07-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mosquito protein AEG12 displays both cytolytic and antiviral properties via a common lipid transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XDJ
 
 | | Crystal Structure Analysis of MBP-SIN3 | | Descriptor: | Maltodextrin-binding protein,Transcriptional regulatory protein SIN3, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of MBP-SIN3
To be published
|
|
2CG8
 
 | | The bifunctional dihydroneopterin aldolase 6-hydroxymethyl-7,8- dihydropterin synthase from Streptococcus pneumoniae | | Descriptor: | DIHYDRONEOPTERIN ALDOLASE 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN SYNTHASE | | Authors: | Garcon, A, Levy, C, Derrick, J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Bifunctional Dihydroneopterin Aldolase/6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Streptococcus Pneumoniae.
J.Mol.Biol., 360, 2006
|
|
