6XH1
 
 | |
8B71
 
 | | Upright KimA dimer with bound c-di-AMP from B. subtilis | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Potassium transporter KimA | | Authors: | Vonck, J, Wieferig, J.P. | | Deposit date: | 2022-09-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic di-AMP traps proton-coupled K + transporters of the KUP family in an inward-occluded conformation.
Nat Commun, 14, 2023
|
|
9CE8
 
 | | 20S Proteasome core particle in complex with Ixazomib | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta, [(1~{R})-1-[2-[[2,5-bis(chloranyl)phenyl]carbonylamino]ethanoylamino]-3-methyl-butyl]boronic acid | | Authors: | Zeytuni, N, Uday, A.B, Vahidi, S, Turner, M. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for allosteric modulation of M. tuberculosis proteasome core particle.
Nat Commun, 16, 2025
|
|
5MJ7
 
 | | Structure of the C. elegans nucleoside hydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Uncharacterized protein | | Authors: | Versees, W, Singh, R.K. | | Deposit date: | 2016-11-30 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the nucleoside hydrolase from C. elegans reveals the role of two active site cysteine residues in catalysis.
Protein Sci., 26, 2017
|
|
8PLZ
 
 | | Cryo-EM structure of CAK in complex with inhibitor CT7030 | | Descriptor: | (3~{R},4~{R})-4-[[[7-[(2-methoxyphenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-06-27 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
7BWV
 
 | |
4ZY7
 
 | |
6Z5A
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941 | | Descriptor: | DIMETHYL SULFOXIDE, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941
To Be Published
|
|
7SXN
 
 | | Orb2A residues 1-9 MYNKFVNFI | | Descriptor: | Orb2A residues 1-9 MYNKFVNFI | | Authors: | Bowler, J.T, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2021-11-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Micro-electron diffraction structure of the aggregation-driving N terminus of Drosophila neuronal protein Orb2A reveals amyloid-like beta-sheets.
J.Biol.Chem., 298, 2022
|
|
6Z7T
 
 | | Nucleotide-free Myosin-II motor domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ewert, W, Preller, M. | | Deposit date: | 2020-06-01 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Computational Insights into a Blebbistatin-Bound Myosin•ADP Complex with Characteristics of an ADP-Release Conformation along the Two-Step Myosin Power Stoke.
Int J Mol Sci, 21, 2020
|
|
6G0G
 
 | |
6RDE
 
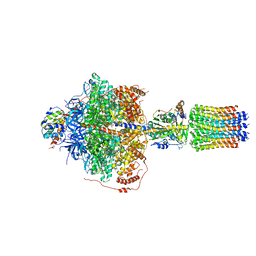 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 2, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE7
 
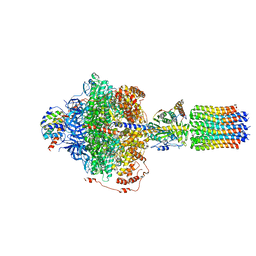 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2C, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9FLC
 
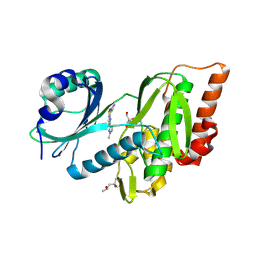 | | Crystal structure of haspin (GSG2) in complex with MU1668 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1-methylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, GLYCEROL, ... | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7T13
 
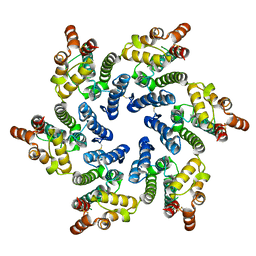 | |
3JZS
 
 | | Human MDM2 liganded with a 12mer peptide inhibitor (pDIQ) | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2, pDIQ peptide (12mer) | | Authors: | Schonbrunn, E, Phan, J. | | Deposit date: | 2009-09-24 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX.
J.Biol.Chem., 285, 2010
|
|
6DFS
 
 | | mouse TCR I.29 in complex with IAg7-p8E9E6ss | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
4RZG
 
 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
6RER
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3B, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9FLO
 
 | | Crystal structure of human Haspin (GSG2) kinase bound to MU2181 | | Descriptor: | NICKEL (II) ION, Serine/threonine-protein kinase haspin, ~{N}-(1,4-dimethylpyrazol-3-yl)-3-(1,2-thiazol-5-yl)thieno[3,2-b]pyridin-5-amine | | Authors: | Kraemer, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6UEC
 
 | | Pseudomonas aeruginosa LpxD Complex Structure with Ligand | | Descriptor: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
8HUK
 
 | | X-ray structure of human PPAR alpha ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUQ
 
 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
6ZB1
 
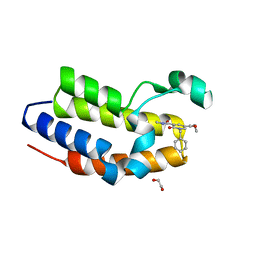 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ~{N}5-cyclopropyl-~{N}3-methyl-2-oxidanylidene-1-(phenylmethyl)pyridine-3,5-dicarboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
9B1H
 
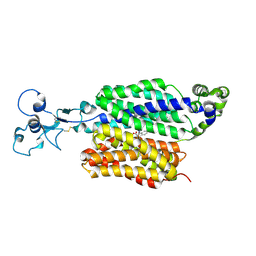 | | Human urate transporter 1 URAT1 in complex with lesinurad | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 22 member 12, lesinurad | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 34, 2024
|
|
