5ED1
 
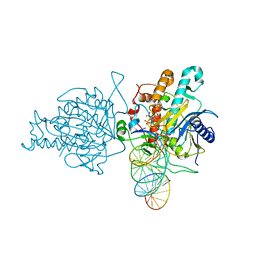 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from S. cerevisiae BDF2 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4BW0
 
 | |
6O5F
 
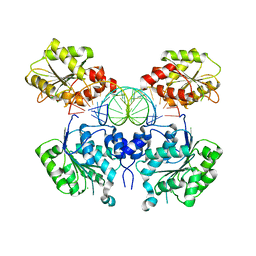 | | Crystal structure of DEAD-box RNA helicase DDX3X at pre-unwound state | | Descriptor: | ATP-dependent RNA helicase DDX3X, CHLORIDE ION, RNA (5'-R(P*CP*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*GP*CP*AP*AP*GP*AP*GP*UP*GP*GP*CP*C)-3') | | Authors: | Song, H, Ji, X. | | Deposit date: | 2019-03-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X.
Nat Commun, 10, 2019
|
|
7S00
 
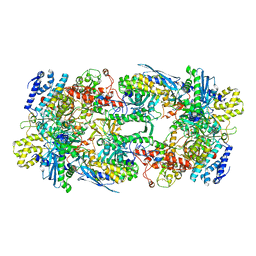 | | X-ray structure of the phage AR9 non-virion RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Sokolova, M.L, Fraser, A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
8XSX
 
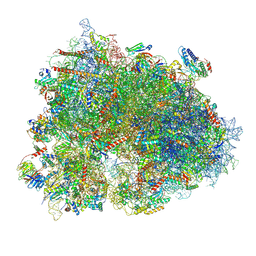 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA, SERBP1 and eEF2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
6BUA
 
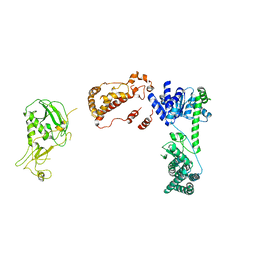 | | Drosophila Dicer-2 apo homology model (helicase, Platform-PAZ, RNaseIII domains) | | Descriptor: | Dicer-2, isoform A | | Authors: | Shen, P.S, Sinha, N.K, Bass, B.L. | | Deposit date: | 2017-12-09 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Dicer uses distinct modules for recognizing dsRNA termini.
Science, 359, 2018
|
|
8ABY
 
 | |
8AD1
 
 | |
8AC0
 
 | |
8ABZ
 
 | |
8ACP
 
 | |
8AC1
 
 | |
5APG
 
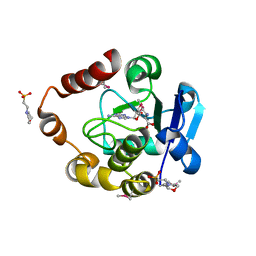 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
8SJ7
 
 | |
7MKN
 
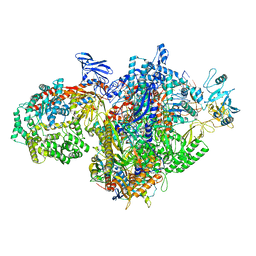 | | Escherichia coli RNA polymerase and RapA elongation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7M8E
 
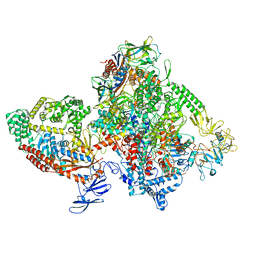 | | E.coli RNAP-RapA elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, W, Liu, B. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
5ELR
 
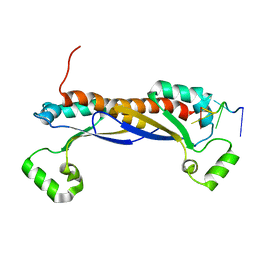 | |
5ELS
 
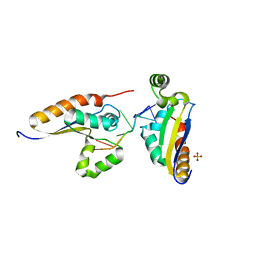 | |
7SLQ
 
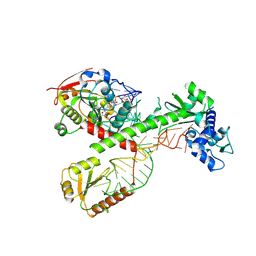 | | Cryo-EM structure of 7SK core RNP with circular RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Minimal circular 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
7SLP
 
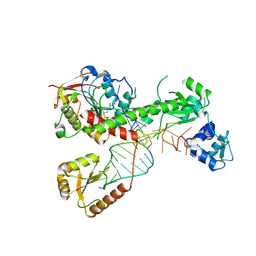 | | Cryo-EM structure of 7SK core RNP with linear RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, La-related protein 7, Linear 7SK RNA, ... | | Authors: | Yang, Y, Liu, S, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-10-24 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP.
Mol.Cell, 82, 2022
|
|
6V5C
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
8DGJ
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGI
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
5WWX
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(P*AP*GP*AP*GP*U)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5WWT
 
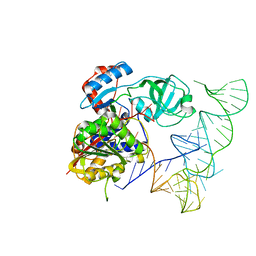 | | Crystal structure of human NSun6/tRNA | | Descriptor: | Putative methyltransferase NSUN6, tRNA | | Authors: | Liu, R.J, Long, T, Wang, E.D. | | Deposit date: | 2017-01-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Nucleic Acids Res., 45, 2017
|
|
