7MI0
 
 | |
5IFK
 
 | | Purine nucleoside phosphorylase | | Descriptor: | HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Thakur, K.G, Priyanka, A. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Functional and Structural Characterization of Purine Nucleoside Phosphorylase from Kluyveromyces lactis and Its Potential Applications in Reducing Purine Content in Food
PLoS ONE, 11, 2016
|
|
8W4Y
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, low-D2O-solvent | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
5IFY
 
 | |
5VJB
 
 | |
7MC6
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8VOU
 
 | | Human glutathione transferase M1-1 in complex with the adduct between glutathione and nitrooleic acid | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1, L-gamma-glutamyl-S-[(8R,9S)-1-carboxy-9-nitroheptadecan-8-yl]-L-cysteinylglycine | | Authors: | Larrieux, N, Steglich, M, Dalla Rizza, J, Buschiazzo, A, Turell, L. | | Deposit date: | 2024-01-16 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Human glutathione transferases catalyze the reaction between glutathione and nitrooleic acid.
J.Biol.Chem., 301, 2025
|
|
5VS1
 
 | |
7MC5
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | Deposit date: | 2021-04-01 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8W4W
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.36 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
6W3Y
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-alanine | | Descriptor: | ALANINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
7MJB
 
 | | Crystal Structure of Nanoluc Luciferase Mutant R164Q | | Descriptor: | CHLORIDE ION, DECANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Reza, M.S, Ai, H, Minor, W. | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Nanoluc Luciferase Mutant R164Q
To Be Published
|
|
5IGS
 
 | | Macrolide 2'-phosphotransferase type I - complex with guanosine and oleandomycin | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, GUANOSINE, ISOPROPYL ALCOHOL, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
6VJ4
 
 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
8VV1
 
 | | Estrogen receptor alpha ligand binding domain in complex with palazestrant | | Descriptor: | Estrogen receptor, GLYCEROL, palazestrant | | Authors: | Ng, R.A, Barratt, S, Parisian, A, Palanisamy, G, Sun, R, Robello, B, Pena, G, Sapugay, J, Yeghikyan, D, Duncan, A, Andersen, S.E, Chawla, R, Rich, B, Hearn, B, Harmon, C, Hodges-Gallagher, L, Kushner, P.J, Greene, G.L, Myles, D.C, Fanning, S.W. | | Deposit date: | 2024-01-30 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Estrogen receptor alpha ligand binding domain in complex with palazestrant
To Be Published
|
|
5VJU
 
 | | De Novo Photosynthetic Reaction Center Protein Variant Equipped with His-Tyr H-bond, Heme B, and Cd(II) ions | | Descriptor: | CADMIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette Leu71His variant | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
6VJE
 
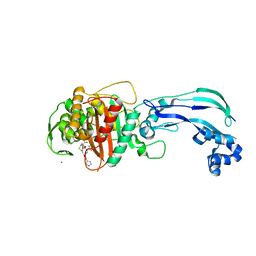 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
8VLD
 
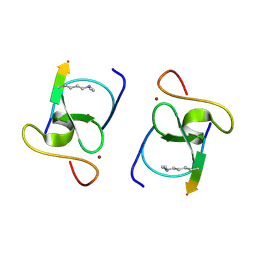 | |
7MDN
 
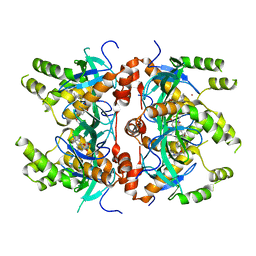 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
8VGS
 
 | |
5VVL
 
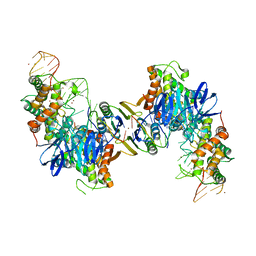 | | Cas1-Cas2 bound to full-site mimic with Ni | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (11-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5VK4
 
 | |
7MJI
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
