2EK0
 
 | |
1VS2
 
 | | Interactions of quinoxaline antibiotic and DNA: the molecular structure of a TRIOSTIN A-D(GCGTACGC) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3', TRIOSTIN A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Rich, A. | | Deposit date: | 1986-10-21 | | Release date: | 2006-06-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions of Quinoxaline Antibiotic and DNA: The Molecular Structure of a Triostin A-D(Gcgtacgc) Complex.
J.Biomol.Struct.Dyn., 4, 1986
|
|
1VT8
 
 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1996-12-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
1VTE
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL A4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1990-05-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc. Natl. Acad. Sci. U.S.A., 87, 1990
|
|
1VCM
 
 | |
2FEZ
 
 | | Mycobacterium tuberculosis EmbR | | Descriptor: | Probable regulatory protein embR | | Authors: | Futterer, K, Alderwick, L.J, Besra, G.S. | | Deposit date: | 2005-12-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F6R
 
 | |
2FIW
 
 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
1VJ4
 
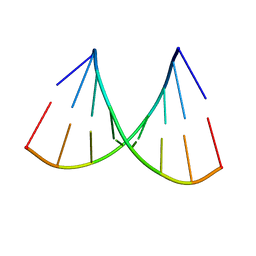 | | SEQUENCE-DEPENDENT CONFORMATION OF AN A-DNA DOUBLE HELIX: THE CRYSTAL STRUCTURE OF THE OCTAMER D(G-G-T-A-T-A-C-C) | | Descriptor: | 5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3' | | Authors: | Shakked, Z, Rabinovich, D, Kennard, O, Cruse, W.B, Salisbury, S.A, Viswamitra, M.A. | | Deposit date: | 1989-01-11 | | Release date: | 1989-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-dependent conformation of an A-DNA double helix: The crystal structure of the octamer d(G-G-T-A-T-A-C-C)
J.Mol.Biol., 166, 1983
|
|
1VJC
 
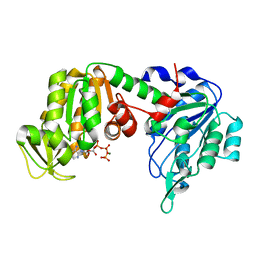 | | Structure of pig muscle PGK complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, phosphoglycerate kinase | | Authors: | Flachner, B, Kovari, Z, Varga, A, Gugolya, Z, Vonderviszt, F, Naray-Szabo, G, Vas, M. | | Deposit date: | 2004-02-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of phosphate chain mobility of MgATP in completing the 3-phosphoglycerate kinase catalytic site: binding, kinetic, and crystallographic studies with ATP and MgATP.
Biochemistry, 43, 2004
|
|
1VQU
 
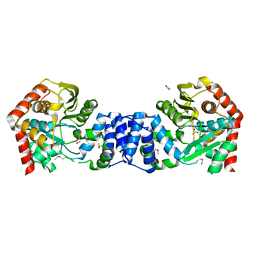 | |
2EYY
 
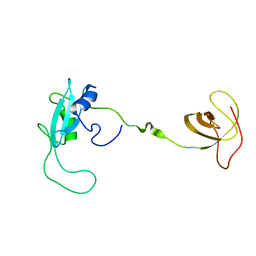 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1VL0
 
 | |
1VPM
 
 | |
1VE3
 
 | | Crystal structure of PH0226 protein from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYLMETHIONINE, hypothetical protein PH0226 | | Authors: | Lokanath, N.K, Yamamoto, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-26 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
2FM9
 
 | |
2F6J
 
 | |
1VTJ
 
 | | MOLECULAR STRUCTURE OF THE NETROPSIN-D(CGCGATATCGCG) COMPLEX: DNA CONFORMATION IN AN ALTERNATING AT SEGMENT; CONFORMATION 1 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Coll, M, Aymami, J, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1999-09-14 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Structure of the Netropsin-d(CGCGATATCGCG) Complex: DNA Conformation in an Alternating AT Segment
Biochemistry, 28, 1989
|
|
1VTV
 
 | | Molecular structure of (M5DC-DG)3: The role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*(CH3)CP*GP*(CH3)CP*G)-3') | | Authors: | Fujii, S, Wang, A.H.-J, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1989-01-10 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Structure of (m5dC-dG)3: The Role of the Methyl Group on 5-Methyl Cytosine in Stabilizing Z-DNA
Nucleic Acids Res., 10, 1982
|
|
2FBD
 
 | | The crystallographic structure of the digestive lysozyme 1 from Musca domestica at 1.90 Ang. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lysozyme 1, SULFATE ION | | Authors: | Cancado, F.C, Marana, S.R, Barbosa, J.A.R.G. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization, data collection and phasing of two digestive lysozymes from Musca domestica.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1VKH
 
 | |
1VT5
 
 | |
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
2DYA
 
 | | Crystal structure of complex between Adenine nucleotide and nucleoside diphosphate kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of complex between Adenine nucleotide and nucleoside diphosphate kinase
To be Published
|
|
1VT7
 
 | |
