8E0P
 
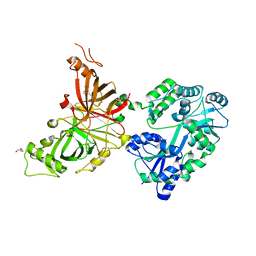 | | Crystal structure of mouse APCDD1 in fusion with engineered MBP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, CHLORIDE ION, ... | | Authors: | Hsieh, F.L, Chang, T.H, Gabelli, S.B, Nathans, J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of WNT inhibitor adenomatosis polyposis coli down-regulated 1 (APCDD1), a cell-surface lipid-binding protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E4H
 
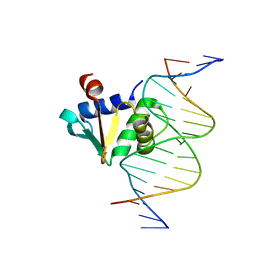 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E3R
 
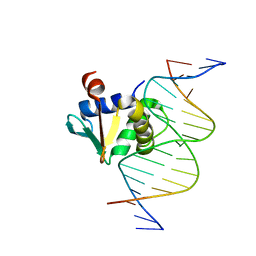 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E3K
 
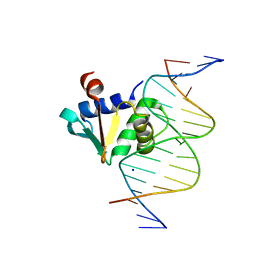 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E5Y
 
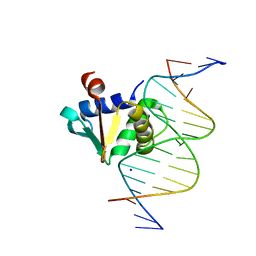 | |
8EE9
 
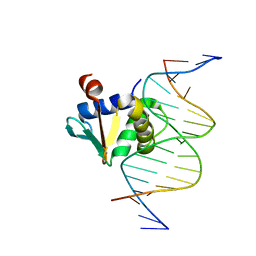 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAATGGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*CP*AP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EBH
 
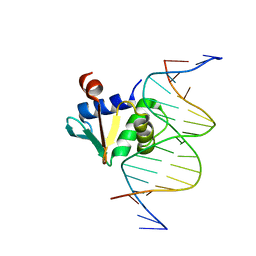 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGAATGGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*CP*AP*TP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-08-31 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8E9A
 
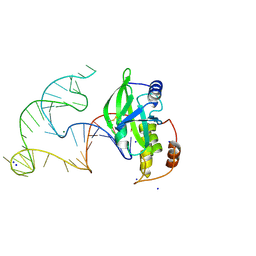 | | Crystal structure of AsfvPolX in complex with 10-23 DNAzyme and Mg | | Descriptor: | DNA/RNA (52-MER), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Cramer, E.R, Robart, A.R, Kaya, A.I. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of a 10-23 deoxyribozyme exhibiting a homodimer conformation.
Commun Chem, 6, 2023
|
|
1SVX
 
 | | Crystal structure of a designed selected Ankyrin Repeat protein in complex with the Maltose Binding Protein | | Descriptor: | Ankyrin Repeat Protein off7, Maltose-binding periplasmic protein | | Authors: | Binz, H.K, Amstutz, P, Kohl, A, Stumpp, M.T, Briand, C, Forrer, P, Gruetter, M.G, Plueckthun, A. | | Deposit date: | 2004-03-30 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | High-affinity binders selected from designed ankyrin repeat protein libraries
NAT.BIOTECHNOL., 22, 2004
|
|
4OGM
 
 | | MBP-fusion protein of PilA1 residues 26-159 | | Descriptor: | Maltose ABC transporter periplasmic protein, pilin protein chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
1ORE
 
 | | Human Adenine Phosphoribosyltransferase | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Thiemann, O.H, Silva, M, Oliva, G, Silva, C.H.T.P, Iulek, J. | | Deposit date: | 2003-03-12 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of human adenine phosphoribosyltransferase and its relation to DHA-urolithiasis.
Biochemistry, 43, 2004
|
|
1TOZ
 
 | | NMR structure of the human NOTCH-1 ligand binding region | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hambleton, S, Valeyev, N.Y, Muranyi, A, Knott, V, Werner, J.M, Mcmichael, A.J, Handford, P.A, Downing, A.K. | | Deposit date: | 2004-06-15 | | Release date: | 2004-10-12 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural and functional properties of the human notch-1 ligand binding region
STRUCTURE, 12, 2004
|
|
1P9Q
 
 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1S7E
 
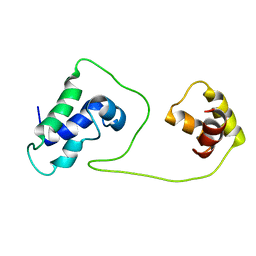 | | Solution structure of HNF-6 | | Descriptor: | Hepatocyte nuclear factor 6 | | Authors: | Liao, X, Sheng, W. | | Deposit date: | 2004-01-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the hepatocyte nuclear factor 6alpha and its interaction with DNA.
J.Biol.Chem., 279, 2004
|
|
1SAV
 
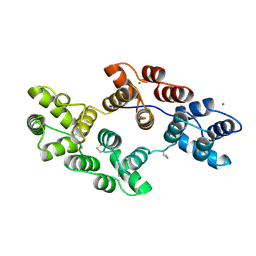 | | HUMAN ANNEXIN V WITH PROLINE SUBSTITUTION BY THIOPROLINE | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Medrano, F.J, Minks, C, Budisa, N, Huber, R. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
1R6Z
 
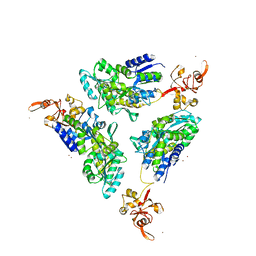 | | The Crystal Structure of the Argonaute2 PAZ domain (as a MBP fusion) | | Descriptor: | Chimera of Maltose-binding periplasmic protein and Argonaute 2, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Song, J.J, Liu, J, Tolia, N.H, Schneiderman, J, Smith, S.K, Martienssen, R.A, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2003-10-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes.
Nat.Struct.Biol., 10, 2003
|
|
1PEB
 
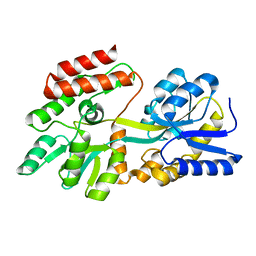 | | LIGAND-FREE HIGH-AFFINITY MALTOSE-BINDING PROTEIN | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
1S3I
 
 | |
4R0Y
 
 | |
1NMU
 
 | | MBP-L30 | | Descriptor: | 60S ribosomal protein L30, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding periplasmic protein | | Authors: | Chao, J.A, Prasad, G.S, White, S.A, Stout, C.D, Williamson, J.R. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inherent Protein Structural Flexibility at the RNA-binding Interface of L30e
J.Mol.Biol., 326, 2003
|
|
4QSZ
 
 | |
1NL5
 
 | | Engineered High-affinity Maltose-Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
4QVH
 
 | | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein | | Descriptor: | CITRATE ANION, COENZYME A, GLYCEROL, ... | | Authors: | Jung, J, Bashiri, G, Johnston, J.M, Baker, E.N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the essential Mycobacterium tuberculosis phosphopantetheinyl transferase PptT, solved as a fusion protein with maltose binding protein.
J.Struct.Biol., 188, 2014
|
|
2K6U
 
 | | The Solution Structure of a Conformationally Restricted Fully Active Derivative of the Human Relaxin-like Factor (RLF) | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-24 | | Release date: | 2008-12-16 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
4RG5
 
 | | Crystal Structure of S. Pombe SMN YG-Dimer | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein, Survival Motor Neuron protein chimera, ... | | Authors: | Gupta, K, Martin, R.S, Sarachan, K.L, Sharp, B, Van Duyne, G.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomeric Properties of Survival Motor NeuronGemin2 Complexes.
J.Biol.Chem., 290, 2015
|
|
