1KVL
 
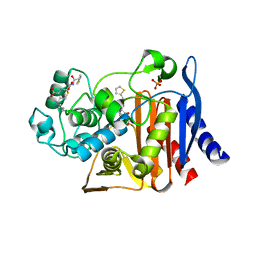 | | X-ray Crystal Structure of AmpC S64G Mutant beta-Lactamase in Complex with Substrate and Product Forms of Cephalothin | | Descriptor: | 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, ... | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
1KVM
 
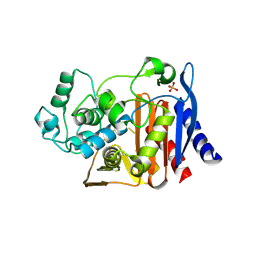 | | X-ray Crystal Structure of AmpC WT beta-Lactamase in Complex with Covalently Bound Cephalothin | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, PHOSPHATE ION, beta-lactamase | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
5U2N
 
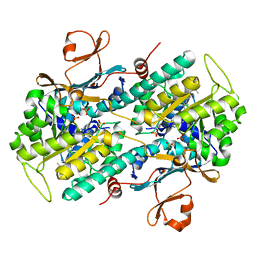 | | Crystal structure of human NAMPT with A-1326133 | | Descriptor: | N-{4-[1-(2-methylpropanoyl)piperidin-4-yl]phenyl}-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
6XJ7
 
 | |
6XJ6
 
 | |
2CLA
 
 | |
4HEF
 
 | | Structure of avibactam bound to Pseudomonas aeruginosa AmpC | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, GLYCEROL | | Authors: | Lahiri, S.D. | | Deposit date: | 2012-10-03 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Insight into Potent Broad-Spectrum Inhibition with Reversible Recyclization Mechanism: Avibactam in Complex with CTX-M-15 and Pseudomonas aeruginosa AmpC beta-Lactamases
Antimicrob.Agents Chemother., 57, 2013
|
|
1GA9
 
 | | CRYSTAL STRUCTURE OF AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH NON-BETA-LACTAMASE INHIBITOR (2, 3-(4-BENZENESULFONYL-THIOPHENE-2-SULFONYLAMINO)-PHENYLBORONIC ACID) | | Descriptor: | 3-(4-BENZENESULFONYL-THIOPHENE-2-SULFONYLAMINO)-PHENYLBORONIC ACID, BETA-LACTAMASE, PHOSPHATE ION, ... | | Authors: | Tondi, D, Powers, R.A, Caselli, E, Negri, M.C, Blazquez, J, Costi, M.P, Shoichet, B.K. | | Deposit date: | 2000-11-29 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design and in-parallel synthesis of inhibitors of AmpC beta-lactamase.
Chem.Biol., 8, 2001
|
|
3TPF
 
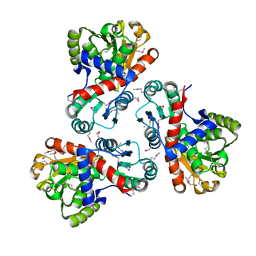 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3KUO
 
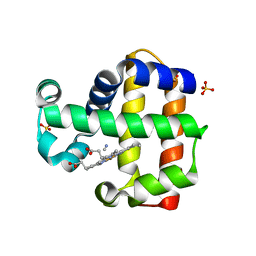 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5UPF
 
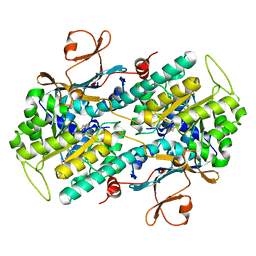 | |
4ETS
 
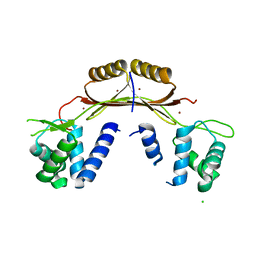 | |
5BJU
 
 | | X-ray structure of the PglF dehydratase from Campylobacter jejuni in complex with UDP and NAD(H) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5UPE
 
 | |
5ADW
 
 | | The Periplasmic Binding Protein CeuE of Campylobacter jejuni preferentially binds the iron(III) complex of the Linear Dimer Component of Enterobactin | | Descriptor: | 2S-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-[(2S)-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-HYDROXY-PROPANOYL]OXY-PROPANOIC ACID, DIMETHYL SULFOXIDE, ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, ... | | Authors: | Raines, D.J, Moroz, O.V, Turkenburg, J.P, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacteria in an Intense Competition for Iron: Key Component of the Campylobacter Jejuni Iron Uptake System Scavenges Enterobactin Hydrolysis Product.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3SWJ
 
 | | Crystal structure of Campylobacter jejuni ChuZ | | Descriptor: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | Authors: | Hu, Y. | | Deposit date: | 2011-07-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
5LQ3
 
 | |
5ADV
 
 | | The Periplasmic Binding Protein CeuE of Campylobacter jejuni preferentially binds the iron(III) complex of the Linear Dimer Component of Enterobactin | | Descriptor: | 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, 2S-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-[(2S)-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-HYDROXY-PROPANOYL]OXY-PROPANOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Raines, D.J, Moroz, O.V, Turkenburg, J.P, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacteria in an Intense Competition for Iron: Key Component of the Campylobacter Jejuni Iron Uptake System Scavenges Enterobactin Hydrolysis Product.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9JQF
 
 | | Crystal structure of the CJ0600 protein from Campylobacter jejuni | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase/D-cysteine desulfhydrase, SULFATE ION | | Authors: | Ki, D.U, Choi, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2024-09-27 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of the CJ0600 protein from Campylobacter jejuni.
Biochem.Biophys.Res.Commun., 735, 2024
|
|
1W2Y
 
 | | The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue dUpNHp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-DIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Galperin, M.Y, Vagin, A.A, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Complex of Campylobacter Jejuni Dutpase with Substrate Analogue Sheds Light on the Mechanism and Suggests the "Basic Module" for Dimeric D(C/U)Tpases
J.Mol.Biol., 342, 2004
|
|
3KUN
 
 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WY4
 
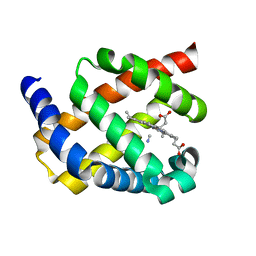 | | Structure of bacterial globin from Campylobacter jejuni at 1.35 A resolution | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SINGLE DOMAIN HAEMOGLOBIN | | Authors: | Barynin, V.V, Sedelnikova, S.E, Shepherd, M, Wu, G, Poole, R.K, Rice, D.W. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Single-Domain Globin from the Pathogenic Bacterium Campylobacter Jejuni: Novel D-Helix Conformation, Proximal Hydrogen Bonding that Influences Ligand Binding, and Peroxidase-Like Redox Properties.
J.Biol.Chem., 285, 2010
|
|
3BSW
 
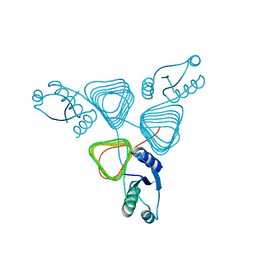 | |
3BSY
 
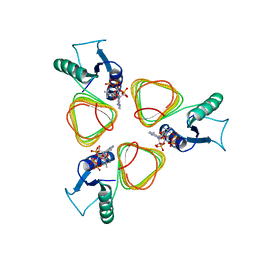 | |
4GIO
 
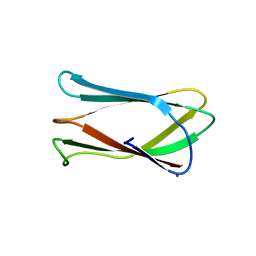 | | Crystal structure of Campylobacter jejuni cj0090 | | Descriptor: | BROMIDE ION, Putative lipoprotein | | Authors: | Kawai, F, Yeo, H.J. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Campylobacter jejuni Cj0090 protein reveals a novel variant of the immunoglobulin fold among bacterial lipoproteins.
Proteins, 80, 2012
|
|
