5CPS
 
 | | Disproportionating enzyme 1 from Arabidopsis - maltotriose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
6L7P
 
 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
5CSY
 
 | | Disproportionating enzyme 1 from Arabidopsis - acarbose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
1AK4
 
 | | HUMAN CYCLOPHILIN A BOUND TO THE AMINO-TERMINAL DOMAIN OF HIV-1 CAPSID | | Descriptor: | CYCLOPHILIN A, HIV-1 CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Vajdos, F.F, Worthylake, D.K, Sundquist, W.I. | | Deposit date: | 1997-05-28 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid.
Cell(Cambridge,Mass.), 87, 1996
|
|
7Z7C
 
 | |
5FOE
 
 | | Crystal structure of the C. elegans Protein O-fucosyltransferase 2 (CePOFUT2) double mutant (R298K-R299K) in complex with GDP and the human TSR1 from thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 2,Thrombospondin-1, ... | | Authors: | Valero-Gonzalez, J, Leonhard-Melief, C, Lira-Navarrete, E, Jimenez-Oses, G, Hernandez-Ruiz, C, Pallares, M.C, Yruela, I, Vasudevan, D, Lostao, A, Corzana, F, Takeuchi, H, Haltiwanger, R.S, Hurtado-Guerrero, R. | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Proactive Role of Water Molecules in Acceptor Recognition of Thrombospondin Type 1 Repeats by Protein-O-Fucosyltransferase 2
Nat.Chem.Biol., 12, 2016
|
|
1B6L
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 4 | | Descriptor: | 1-[2-(8-CARBAMOYLMETHYL-6,9-DIOXO-2-OXA-7,10-DIAZA-BICYCLO[11.2.2]HEPTADECA- 1(16),13(17),14-TRIEN-11-YL)-2-HYDROXY-ETHYL]-PIPERIDINE-2-CARBOXYLIC ACID TERT-BUTYLAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
4O0X
 
 | | Back pocket flexibility provides group-II PAK selectivity for type 1 kinase inhibitors | | Descriptor: | 1-{[1-(4-amino-1,3,5-triazin-2-yl)-2-methyl-1H-benzimidazol-6-yl]ethynyl}cyclohexanol, Serine/threonine-protein kinase PAK 4 | | Authors: | Rouge, L, Tam, C, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Back Pocket Flexibility Provides Group II p21-Activated Kinase (PAK) Selectivity for Type I 1/2 Kinase Inhibitors.
J.Med.Chem., 57, 2014
|
|
5X7R
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
6C89
 
 | | NDM-1 Beta-Lactamase Exhibits Differential Active Site Sequence Requirements for the Hydrolysis of Penicillin versus Carbapenem Antibiotics | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Palzkill, T, Sun, Z, Sankaran, B. | | Deposit date: | 2018-01-24 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75006151 Å) | | Cite: | Differential active site requirements for NDM-1 beta-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics.
Nat Commun, 9, 2018
|
|
6HQ6
 
 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Bacterial beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
7WJB
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJA
 
 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P6322 space group | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7Y3S
 
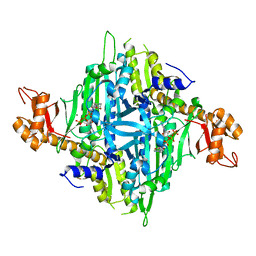 | | Controlling fibrosis using compound with novel binding mode to prolyl-tRNA synthetase 1 | | Descriptor: | 1-(6-bromanyl-7-methyl-imidazo[4,5-b]pyridin-3-yl)-3-[(2R,3S)-3-oxidanylpiperidin-2-yl]propan-2-one, ADENOSINE-5'-TRIPHOSPHATE, Bifunctional glutamate/proline--tRNA ligase, ... | | Authors: | Kim, S, Yoon, I, Son, J, Park, S, Hwang, K.Y. | | Deposit date: | 2022-06-12 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Control of fibrosis with enhanced safety via asymmetric inhibition of prolyl-tRNA synthetase 1.
Embo Mol Med, 15, 2023
|
|
7WCE
 
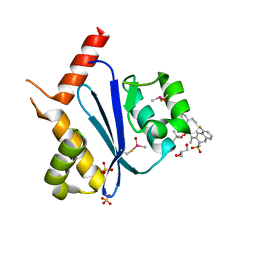 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with (2S)-2-(tert-Butoxy)-2-(10-fluoro-2-(2-hydroxy-4-methylphenyl)-1,4-dimethyl-5-(methylsulfonyl)-5,6-dihydrophenanthridin-3-yl)acetic acid | | Descriptor: | (2S)-2-[10-fluoranyl-1,4-dimethyl-2-(4-methyl-2-oxidanyl-phenyl)-5-methylsulfonyl-6H-phenanthridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, GLYCEROL, Integrase catalytic, ... | | Authors: | Taoda, Y, Sekiguchi, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of tricyclic HIV-1 integrase-LEDGF/p75 allosteric inhibitors by intramolecular direct arylation reaction.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
2VG7
 
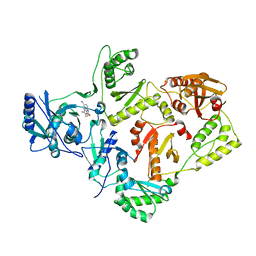 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-iodophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
6KJL
 
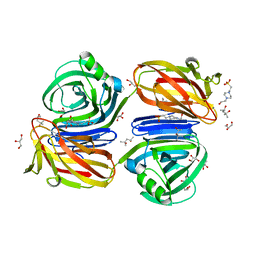 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, M, Takita, T, Nakatani, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
4B3Q
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, P51 RT, PRIMER DNA, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
6ALD
 
 | | RABBIT MUSCLE ALDOLASE A/FRUCTOSE-1,6-BISPHOSPHATE COMPLEX | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), FRUCTOSE-1,6-BIS(PHOSPHATE) ALDOLASE | | Authors: | Choi, K.H, Mazurkie, A.S, Morris, A.J, Utheza, D, Tolan, D.R, Allen, K.N. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 A(,).
Biochemistry, 38, 1999
|
|
4B3O
 
 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
6B38
 
 | |
6B3G
 
 | |
6UL5
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
4O0V
 
 | |
