1PIK
 
 | | ESPERAMICIN A1-DNA COMPLEX, NMR, 4 STRUCTURES | | Descriptor: | 2,4-dideoxy-3-O-methyl-4-(propan-2-ylamino)-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-S-methyl-4-thio-beta-D-ribo-hexopyranose, 2-deoxy-alpha-L-fucopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Esperamicin A1-DNA Complex
J.Mol.Biol., 265, 1997
|
|
1PBW
 
 | | STRUCTURE OF BCR-HOMOLOGY (BH) DOMAIN | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Musacchio, A, Cantley, L.C, Harrison, S.C. | | Deposit date: | 1996-10-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1TOM
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE | | Authors: | Chen, Z. | | Deposit date: | 1996-12-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, evaluation, and crystallographic analysis of L-371,912: A potent and selective active-site thrombin inhibitor.
Bioorg.Med.Chem.Lett., 7, 1997
|
|
1DJA
 
 | |
1FKZ
 
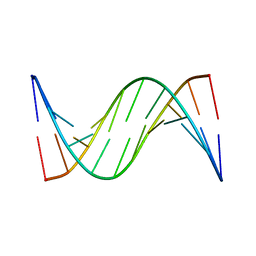 | | NMR STUDY OF B-DNA CONTAINING A MISMATCHED BASE PAIR IN THE 29-39 K-RAS GENE SEQUENCE: CC CT C+C C+T, 2 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*CP*AP*GP*CP*TP*C)-3') | | Authors: | Boulard, Y, Cognet, J.A.H, Fazakerley, G.V. | | Deposit date: | 1996-10-09 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure as a function of pH of two central mismatches, C . T and C . C, in the 29 to 39 K-ras gene sequence, by nuclear magnetic resonance and molecular dynamics.
J.Mol.Biol., 268, 1997
|
|
1SEI
 
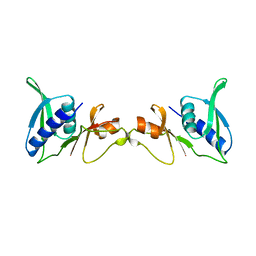 | | STRUCTURE OF 30S RIBOSOMAL PROTEIN S8 | | Descriptor: | RIBOSOMAL PROTEIN S8 | | Authors: | Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-08-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence for specific S8-RNA and S8-protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 A resolution.
Structure, 4, 1996
|
|
1FOX
 
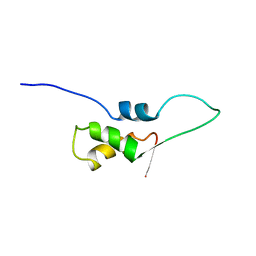 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1HSE
 
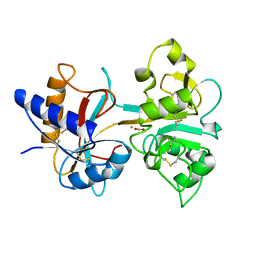 | | H253M N TERMINAL LOBE OF HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Nicholson, H, Anderson, B.F, Baker, E.N. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis of the histidine ligand in human lactoferrin: iron binding properties and crystal structure of the histidine-253-->methionine mutant.
Biochemistry, 36, 1997
|
|
1ONR
 
 | | STRUCTURE OF TRANSALDOLASE B | | Descriptor: | TRANSALDOLASE B | | Authors: | Jia, J, Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-08-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of transaldolase B from Escherichia coli suggests a circular permutation of the alpha/beta barrel within the class I aldolase family.
Structure, 4, 1996
|
|
1JHG
 
 | | TRP REPRESSOR MUTANT V58I | | Descriptor: | PHOSPHATE ION, TRP OPERON REPRESSOR, TRYPTOPHAN | | Authors: | Lawson, C.L. | | Deposit date: | 1996-07-19 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An atomic view of the L-tryptophan binding site of trp repressor.
Nat.Struct.Biol., 3, 1996
|
|
1HKL
 
 | | FREE AND LIGANDED FORM OF AN ESTEROLYTIC CATALYTIC ANTIBODY | | Descriptor: | 48G7 FAB | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-03-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structures of the free and liganded form of an esterolytic catalytic antibody.
J.Mol.Biol., 268, 1997
|
|
1HRD
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
1FOW
 
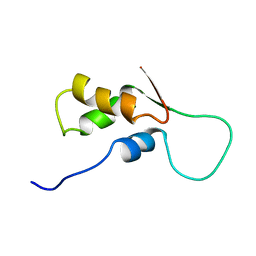 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1HPJ
 
 | |
1RC4
 
 | |
1VJS
 
 | |
1AII
 
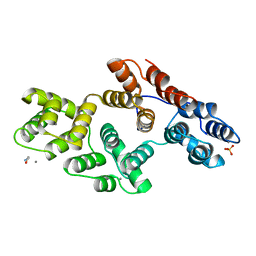 | | ANNEXIN III | | Descriptor: | ANNEXIN III, CALCIUM ION, ETHANOLAMINE, ... | | Authors: | Lewit-Bentley, A, Perron, B. | | Deposit date: | 1996-11-28 | | Release date: | 1997-03-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Can enzymatic activity, or otherwise, be inferred from structural studies of annexin III?
J.Biol.Chem., 272, 1997
|
|
1YTG
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YFC
 
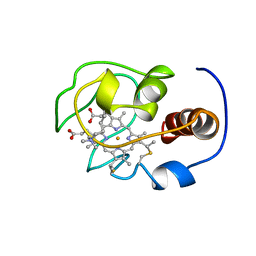 | | Solution nmr structure of a yeast iso-1-ferrocytochrome C | | Descriptor: | HEME C, YEAST ISO-1-FERROCYTOCHROME C | | Authors: | Baistrocchi, P, Banci, L, Bertini, I, Turano, P, Bren, K.L, Gray, H.B. | | Deposit date: | 1996-08-08 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Saccharomyces cerevisiae reduced iso-1-cytochrome c.
Biochemistry, 35, 1996
|
|
1YTJ
 
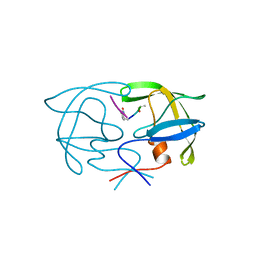 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | PEPTIDE PRODUCT, SIV PROTEASE | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YTH
 
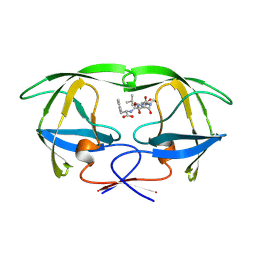 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
2BNH
 
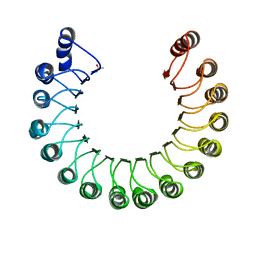 | | PORCINE RIBONUCLEASE INHIBITOR | | Descriptor: | RIBONUCLEASE INHIBITOR | | Authors: | Kobe, B, Deisenhofer, J. | | Deposit date: | 1996-06-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A.
J.Mol.Biol., 264, 1996
|
|
3CEL
 
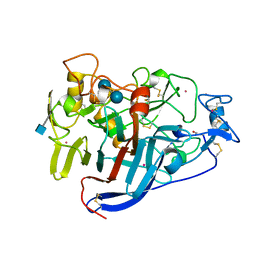 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1UCB
 
 | |
1NOX
 
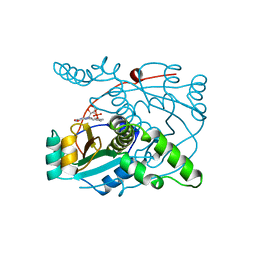 | | NADH OXIDASE FROM THERMUS THERMOPHILUS | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH OXIDASE | | Authors: | Hecht, H.J, Erdmann, H, Park, H.J, Sprinzl, M, Schmid, R.D. | | Deposit date: | 1996-11-20 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of NADH oxidase from Thermus thermophilus.
Nat.Struct.Biol., 2, 1995
|
|
