6O7V
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7OI7
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 2 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
6OA8
 
 | | Superfolder Green Fluorescent Protein with 4-cyano-L-phenylalanine at the chromophore (position 66) | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION, ... | | Authors: | Piacentini, J, Olenginski, G.M, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
7Q4G
 
 | | Structure of coproheme decarboxylase from Corynebacterium dipththeriae Y135A mutant in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Coproheme decarboxylase from Corynebacterium diphtheriae Y135A mutant in complex with coproheme, DI(HYDROXYETHYL)ETHER | | Authors: | Michlits, H, Valente, N, Mlynek, G, Hofbauer, S. | | Deposit date: | 2021-10-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Initial Steps to Engineer Coproheme Decarboxylase to Obtain Stereospecific Monovinyl, Monopropionyl Deuterohemes.
Front Bioeng Biotechnol, 9, 2021
|
|
6WKD
 
 | | Crystal structure of pentalenene synthase complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
7OIC
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 4 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OI8
 
 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 3A | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
5WJO
 
 | | Crystal structure of the unliganded PG90 TCR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PG90 TCR alpha chain, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
4XR8
 
 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
6OFY
 
 | | Crystal Structure of Arachidonic Acid bound to V349I murine COX-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, ARACHIDONIC ACID, ... | | Authors: | Malkowski, M.G. | | Deposit date: | 2019-04-01 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arg-513 and Leu-531 Are Key Residues Governing Time-Dependent Inhibition of Cyclooxygenase-2 by Aspirin and Celebrex.
Biochemistry, 58, 2019
|
|
6NUN
 
 | |
7OID
 
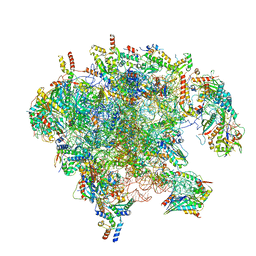 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 5A | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
9MFR
 
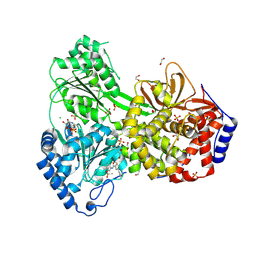 | | Cat DHX9 in Complex with Compound 23 and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Lockbaum, G.J, Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A, Grigoriu, S. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of ATX968: An Orally Available Allosteric Inhibitor of DHX9.
J.Med.Chem., 68, 2025
|
|
6NLX
 
 | |
6ORJ
 
 | |
5O08
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with Dephospho-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
8EO5
 
 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
7PWC
 
 | | PARP15 catalytic domain in complex with OUL238 | | Descriptor: | 4-(cyclobutylmethoxy)benzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
6OII
 
 | |
4G4M
 
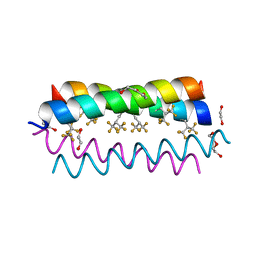 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3(6-13) | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, alpha4F3(6-13) | | Authors: | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2012-07-16 | | Release date: | 2012-10-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison of the structures and stabilities of coiled-coil proteins containing hexafluoroleucine and t-butylalanine provides insight into the stabilizing effects of highly fluorinated amino acid side-chains.
Protein Sci., 21, 2012
|
|
7PWA
 
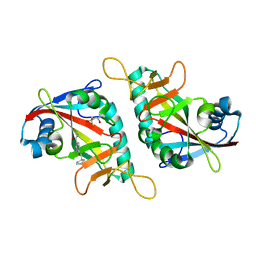 | | PARP15 catalytic domain in complex with OUL237 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(cyclopentylmethoxy)benzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PW3
 
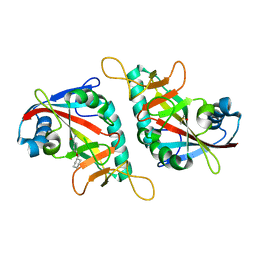 | | PARP15 catalytic domain in complex with OUL217 | | Descriptor: | 4-(cyclohexylmethoxy)benzamide, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
8GR6
 
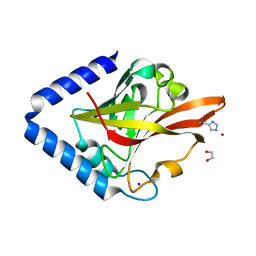 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
6WLX
 
 | |
7PWK
 
 | | PARP15 catalytic domain in complex with OUL239 | | Descriptor: | 4-(cyclopropylmethoxy)benzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
