6UPP
 
 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3IXY
 
 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
5EAG
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor Prochloraz | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Prochloraz | | Authors: | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
5O3G
 
 | | Human Brd2(BD2) mutant in complex with AL-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-pent-4-enamide, Bromodomain-containing protein 2 | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
5HHJ
 
 | |
5EAD
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor S-desthio-prothioconazole | | Descriptor: | (2~{S})-2-(1-chloranylcyclopropyl)-1-(2-chlorophenyl)-3-(1,2,4-triazol-1-yl)propan-2-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tyndall, J.D.A, Sabherwal, M, Sagatova, A.A, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
7ZYH
 
 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 4, Isoform 4 of Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Muckenfuss, L.M, Jinek, M, Migenda Herranz, A.C, Clerici, M. | | Deposit date: | 2022-05-24 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fip1 is a multivalent interaction scaffold for processing factors in human mRNA 3' end biogenesis.
Elife, 11, 2022
|
|
1AOB
 
 | | E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH DDURD | | Descriptor: | 2'-5'DIDEOXYURIDINE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
5HL9
 
 | | E. coli PBP1b in complex with acyl-ampicillin and moenomycin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into Inhibition of Escherichia coli Penicillin-binding Protein 1B.
J.Biol.Chem., 292, 2017
|
|
5HLH
 
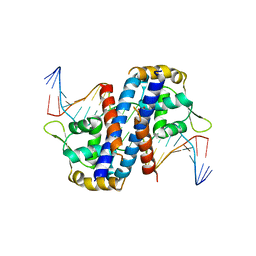 | | Crystal structure of the overoxidized AbfR bound to DNA | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*TP*CP*AP*AP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*TP*GP*AP*GP*T)-3'), MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.-G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
5DSP
 
 | |
2XCH
 
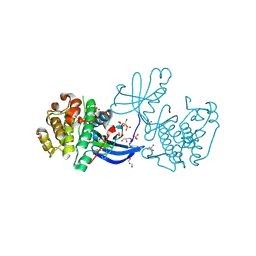 | | Crystal structure of PDK1 in complex with a pyrazoloquinazoline inhibitor | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, 8-(CYCLOHEXA-2,5-DIEN-1-YLIDENEAMINO)-1-(PIPERIDIN-4-YLMETHYL)-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, GLYCEROL, ... | | Authors: | Angiolini, M, Banfi, P, Casale, E, Casuscelli, F, Fiorelli, C, Saccardo, M.B, Silvagni, M, Zuccotto, F. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Optimization of Potent Pdk1 Inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6UWX
 
 | |
3IIZ
 
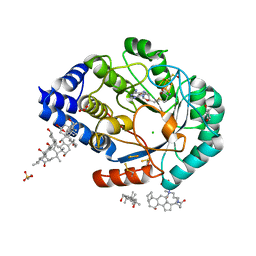 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with S-adenosyl-L-methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
8D9O
 
 | | De Novo Photosynthetic Reaction Center Protein in Apo-State | | Descriptor: | CADMIUM ION, Reaction Center Maquette | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
4RXR
 
 | | The structure of GTP-dCTP-bound SAMHD1 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhu, C.F, Wei, W, Peng, X, Dong, Y.H, Gong, Y, Yu, X.F. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ICJ
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus furiosus | | Descriptor: | ZINC ION, uncharacterized metal-dependent hydrolase | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Furiosus
To be Published
|
|
2HEH
 
 | | Crystal Structure of the KIF2C motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF2C protein, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the kif2c motor domain
to be published
|
|
4RU1
 
 | | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus 11B, TARGET EFI-510965, in complex with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CITRIC ACID, Monosaccharide ABC transporter substrate-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of carbohydrate transporter ACEI_1806 from Acidothermus cellulolyticus, TARGET EFI-510965.
To be Published
|
|
2HCM
 
 | | Crystal structure of mouse putative dual specificity phosphatase complexed with zinc tungstate, New York Structural Genomics Consortium | | Descriptor: | Dual specificity protein phosphatase, GLYCEROL, SODIUM ION, ... | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1BDU
 
 | |
6UQG
 
 | | Human HCN1 channel Y289D mutant | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
2N4F
 
 | | EC-NMR Structure of Arabidopsis thaliana At2g32350 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein AR3433A | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2BZS
 
 | | Binding of anti-cancer prodrug CB1954 to the activating enzyme NQO2 revealed by the crystal structure of their complex. | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH DEHYDROGENASE [QUINONE] 2, ... | | Authors: | Abu Khader, M.M, Heap, J.T, De Matteis, C, Kellam, B, Doughty, S.W, Minton, N, Paoli, M. | | Deposit date: | 2005-08-22 | | Release date: | 2005-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of the Anticancer Prodrug Cb1954 to the Activating Enzyme Nqo2 Revealed by the Crystal Structure of Their Complex.
J.Med.Chem., 48, 2005
|
|
2XM9
 
 | | Structure of a small molecule inhibitor with the kinase domain of Chk2 | | Descriptor: | 4-(1H-pyrazol-5-yl)-2-{4-[(3S)-pyrrolidin-3-ylamino]quinazolin-2-yl}phenol, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Caldwell, J.J, Welsh, E.J, Matijssen, C, Anderson, V.E, Antoni, L, Boxall, K, Urban, F, Hayes, A, Raynaud, F.I, Rigoreau, L.J, Raynham, T, Aherne, G.W, Pearl, L.H, Oliver, A.W, Garrett, M.D, Collins, I. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Potent and Selective 2-(Quinazolin-2-Yl)Phenol Inhibitors of Checkpoint Kinase 2.
J.Med.Chem., 54, 2011
|
|
