3M7T
 
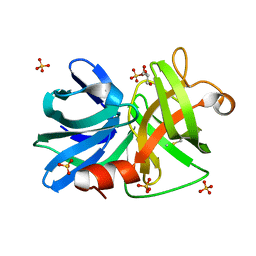 | |
7A61
 
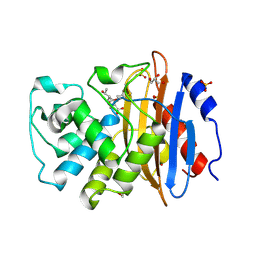 | | Crystal structure of KPC-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (2~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-butyl-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A63
 
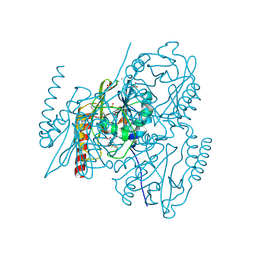 | | Crystal structure of L1 with hydrolyzed faropenem (imine, ring-closed form) | | Descriptor: | (2R,5S)-2-[(1S,2R)-1-carboxy-2-hydroxy-propyl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydrothiazole-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57000113 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
1M7G
 
 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
3UPF
 
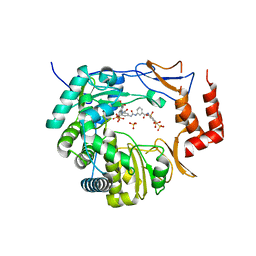 | | Crystal structure of murine norovirus RNA-dependent RNA polymerase bound to NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
9G0G
 
 | | BsCdaA in complex with Compound 7 | | Descriptor: | CHLORIDE ION, Cyclic di-AMP synthase CdaA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8IND
 
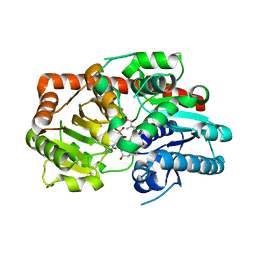 | | Crystal structure of UGT74AN3-UDP-RES | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | Authors: | Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
9GL9
 
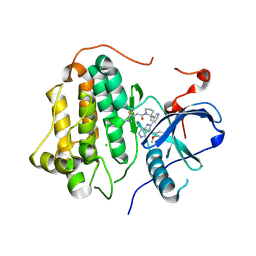 | | Wild-type EGFR bound with STX-721 | | Descriptor: | (2R,3S)-3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[2-[(2R)-1-[(E)-4-(dimethylamino)but-2-enoyl]-2-methyl-pyrrolidin-2-yl]ethynyl]pyridin-4-yl]-1,2,3,5,6,7-hexahydropyrrolo[3,2-c]pyridin-4-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 31, 2025
|
|
8INV
 
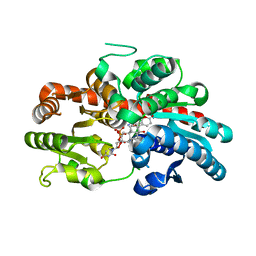 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
6DAL
 
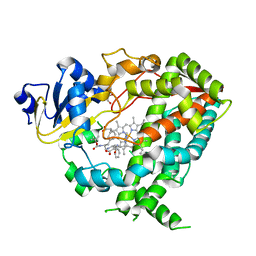 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, Nalpha-{(2S)-2-[(tert-butoxycarbonyl)amino]-3-phenylpropyl}-N-[2-(pyridin-3-yl)ethyl]-D-phenylalaninamide, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
3E88
 
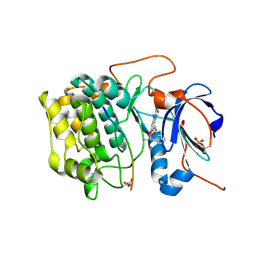 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3UT6
 
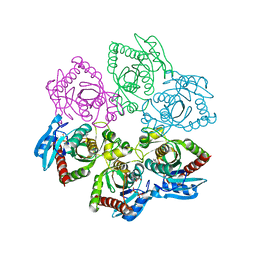 | | Crystal structure of E. Coli PNP complexed with PO4 and formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Stefanic, Z. | | Deposit date: | 2011-11-25 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | New phosphate binding sites in the crystal structure of Escherichia coli purine nucleoside phosphorylase complexed with phosphate and formycin A.
Febs Lett., 586, 2012
|
|
8Q58
 
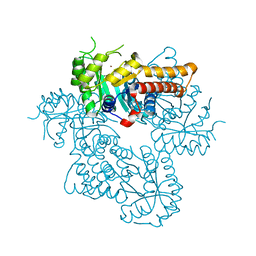 | |
3MR0
 
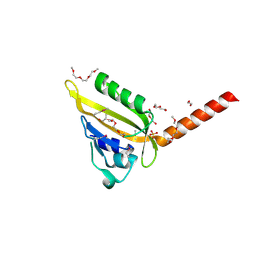 | | Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264
To be Published
|
|
1ME4
 
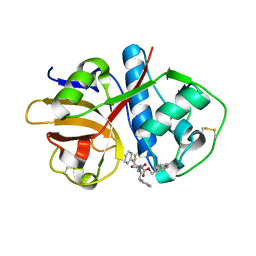 | |
3V1R
 
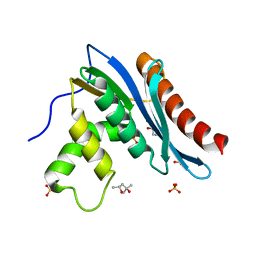 | | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of XMRV with inhibitor beta-thujaplicinol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, ... | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the reverse transcriptase-associated ribonuclease H domain of xenotropic murine leukemia-virus related virus.
J.Struct.Biol., 177, 2012
|
|
5F6X
 
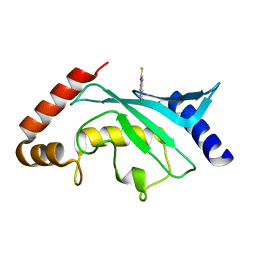 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole from cocktail screen) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8INO
 
 | | Crystal structure of UGT74AN3 in complex UDP and PER | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(3S,5S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3,5,14-tris(oxidanyl)-2,3,4,6,7,8,9,11,12,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2H-furan-5-one, Glycosyltransferase, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
9G25
 
 | | snR30 snoRNP - State 1 - Utp23-Krr1-deltaC3 | | Descriptor: | 40S ribosomal protein S13, 40S ribosomal protein S14-A, H/ACA ribonucleoprotein complex subunit CBF5, ... | | Authors: | Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.
Nat Commun, 16, 2025
|
|
6SWO
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 2 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2PCO
 
 | | Spatial Structure and Membrane Permeabilization for Latarcin-1, a Spider Antimicrobial Peptide | | Descriptor: | Latarcin-1 | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity.
Biochemistry, 47, 2008
|
|
9G5H
 
 | | p53-Y220C Core Domain Covalently Bound to 2-chloro-5-cyanopyrazine Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranylpyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-17 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
9G6U
 
 | | p53-Y220C Core Domain Covalently Bound to 3,5-Dichloro-6-Ethylpyrazine-2-carbonitirle Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(chloranyl)-6-ethyl-pyrazine-2-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | S N Ar Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes.
Drug Des Devel Ther, 19, 2025
|
|
3UNX
 
 | | Bond length analysis of asp, glu and his residues in subtilisin Carlsberg at 1.26A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Fisher, S.J, Helliwell, J.R, Blakeley, M.P, Cianci, M, McSweeny, S. | | Deposit date: | 2011-11-16 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Protonation-state determination in proteins using high-resolution X-ray crystallography: effects of resolution and completeness.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
