5LWM
 
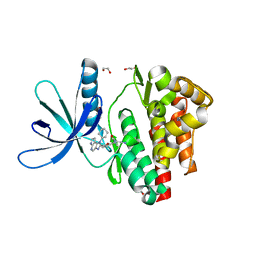 | | Crystal structure of JAK3 in complex with Compound 4 (FM381) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 2-cyano-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl)furan-2-yl]-~{N},~{N}-dimethyl-prop-2-enamide, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
8IOB
 
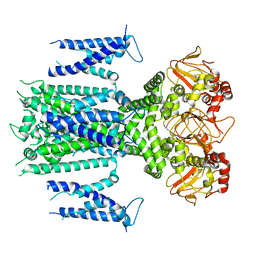 | | Herg1a-herg1b closed state 1 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Herg1a-herg1b closed state 1
To Be Published
|
|
8I42
 
 | |
9GRG
 
 | | StmPr1, Stenotrophomonas maltophilia Protease 1, 36 kDa alkine serine protease in complex with PMSF | | Descriptor: | Alkaline serine protease, CALCIUM ION, GLYCEROL, ... | | Authors: | Sommer, M, Outzen, L, Negm, A, Windhorst, S, Weber, W, Betzel, C. | | Deposit date: | 2024-09-11 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep, 15, 2025
|
|
1M9L
 
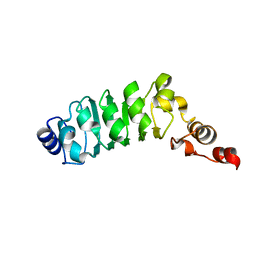 | | Relaxation-based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Outer Arm Dynein Light Chain 1 | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relaxation-based structure refinement and backbone molecular dynamics of the
Dynein motor domain-associated light chain
Biochemistry, 42, 2003
|
|
8I47
 
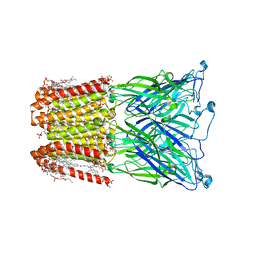 | |
8INU
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
6SW8
 
 | | Crystal structure of the NS1 (H7N1) RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural protein 1 | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
3EAM
 
 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
6ZVA
 
 | | Crystal structure of My5 | | Descriptor: | Myomesin-1, SULFATE ION | | Authors: | Sauer, F, Wilmanns, M. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Myomesin-1
To Be Published
|
|
6ZW1
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | Descriptor: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | Authors: | Barinka, C, Motlova, L, Ustinova, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
6ZXI
 
 | | Crystal Structure of the OXA-48 Carbapenem-Hydrolyzing Class D beta-Lactamase in Complex with the DBO inhibitor ANT3310 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ANT3310 , a Novel Broad-Spectrum Serine beta-Lactamase Inhibitor of the Diazabicyclooctane Class, Which Strongly Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacterales and Acinetobacter baumannii.
J.Med.Chem., 63, 2020
|
|
3VGP
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AF_0329) from Archaeoglobus fulgidus | | Descriptor: | Transmembrane oligosaccharyl transferase, putative | | Authors: | Matsumoto, S, Igura, M, Nyirenda, J, Yuzawa, S, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the C-Terminal Globular Domain of Oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 A Resolution
Biochemistry, 51, 2012
|
|
3MVE
 
 | | Crystal structure of a novel pyruvate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UPF0255 protein VV1_0328 | | Authors: | Cha, S.S, Jeong, C.S, An, Y.J. | | Deposit date: | 2010-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | FrsA functions as a cofactor-independent decarboxylase to control metabolic flux.
Nat.Chem.Biol., 7, 2011
|
|
6SQZ
 
 | | Mouse dCTPase in complex with dCMPNPP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
8IZC
 
 | | Human CK1 Delta Kinase structure bound to Inhibitor | | Descriptor: | Casein kinase I isoform delta, SULFATE ION, ~{N}5-~{tert}-butyl-2-(3-chloranyl-4-fluoranyl-phenyl)-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-3,5-dicarboxamide | | Authors: | Ghosh, K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Discovery of BMS-986164, a Potent, Selective and Orally Efficacious CK1d/e/a Inhibitor from Pyrazolo-Piperazine Chemotype
To Be Published
|
|
8Q29
 
 | |
1MMD
 
 | | TRUNCATED HEAD OF MYOSIN FROM DICTYOSTELIUM DISCOIDEUM COMPLEXED WITH MGADP-BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Holden, H.M, Rayment, I. | | Deposit date: | 1995-03-21 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4-.
Biochemistry, 34, 1995
|
|
6D8J
 
 | |
3VND
 
 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
7A21
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2158 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type I, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-08-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2153
To Be Published
|
|
1DVA
 
 | |
3UUO
 
 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1M0U
 
 | | Crystal Structure of the Drosophila Glutathione S-transferase-2 in Complex with Glutathione | | Descriptor: | GLUTATHIONE, GST2 gene product, SULFATE ION | | Authors: | Agianian, B, Tucker, P.A, Schouten, A, Leonard, K, Bullard, B, Gros, P. | | Deposit date: | 2002-06-14 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Drosophila Sigma Class Glutathione S-transferase Reveals a Novel
Active Site Topography Suited for Lipid Peroxidation Products
J.Mol.Biol., 326, 2003
|
|
3E8Z
 
 | |
