2IGW
 
 | | CYCLOPHILIN 3 complexed with DIPEPTIDE GLY-PRO | | Descriptor: | GLYCINE, PROLINE, Peptidyl-prolyl cis-trans isomerase 3 | | Authors: | Kan, D. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Experimental Determination of van der Waals Energies in a Biological System.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2IGX
 
 | | Achiral, Cheap and Potent Inhibitors of Plasmepsins II | | Descriptor: | 5-PENTYL-N-{[4'-(PIPERIDIN-1-YLCARBONYL)BIPHENYL-4-YL]METHYL}-N-[1-(PYRIDIN-2-YLMETHYL)PIPERIDIN-4-YL]PYRIDINE-2-CARBOXAMIDE, Plasmepsin-2 | | Authors: | Prade, L. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-28 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Achiral, Cheap, and Potent Inhibitors of Plasmepsins I, II, and IV.
Chemmedchem, 1, 2006
|
|
2IGY
 
 | | Achiral, Cheap and Potent Inhibitors of Plasmepsins II | | Descriptor: | N-[1-(3-METHYLBUTYL)PIPERIDIN-4-YL]-N-{4-[METHYL(PYRIDIN-4-YL)AMINO]BENZYL}-4-PENTYLBENZAMIDE, Plasmepsin-2 | | Authors: | Prade, L. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Achiral, Cheap, and Potent Inhibitors of Plasmepsins I, II, and IV.
Chemmedchem, 1, 2006
|
|
2IGZ
 
 | |
2IH0
 
 | |
2IH1
 
 | | Ion selectivity in a semi-synthetic K+ channel locked in the conductive conformation | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB Heavy Chain, FAB Light Chain, ... | | Authors: | Valiyaveetil, F.I, Leonetti, M, Muir, T.W, MacKinnon, R. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ion Selectivity in a Semisynthetic K+ Channel Locked in the Conductive Conformation.
Science, 314, 2006
|
|
2IH2
 
 | |
2IH3
 
 | | Ion selectivity in a semi-synthetic K+ channel locked in the conductive conformation | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB Heavy Chain, FAB Light Chain, ... | | Authors: | Valiyaveetil, F.I, Leonetti, M, Muir, T.W, MacKinnon, R. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ion Selectivity in a Semisynthetic K+ Channel Locked in the Conductive Conformation.
Science, 314, 2006
|
|
2IH4
 
 | |
2IH5
 
 | |
2IH6
 
 | | Pro6 variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IH7
 
 | | Amidated Pro6 Analogue of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IH8
 
 | | A low-dose crystal structure of a recombinant Melanocarpus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A crystallographic and spectroscopic study on the effect of X-ray radiation on the crystal structure of Melanocarpus albomyces laccase.
Biochem.Biophys.Res.Commun., 350, 2006
|
|
2IH9
 
 | | A high-dose crystal structure of a recombinant Melanocarbus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A crystallographic and spectroscopic study on the effect of X-ray radiation on the crystal structure of Melanocarpus albomyces laccase.
Biochem.Biophys.Res.Commun., 350, 2006
|
|
2IHA
 
 | | Amidated variant of CMrVIA conotoxin | | Descriptor: | Lambda-conotoxin CMrVIA | | Authors: | Kini, R.M, Kang, T.S. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-14 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins
Biochemistry, 46, 2007
|
|
2IHB
 
 | | Crystal structure of the heterodimeric complex of human RGS10 and activated Gi alpha 3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, MAGNESIUM ION, ... | | Authors: | Soundararajan, M, Turnbull, A.P, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Ugochukwu, E, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2IHC
 
 | | Crystal structure of the bric-a-brac (BTB) domain of human BACH1 | | Descriptor: | Transcription regulator protein BACH1 | | Authors: | Amos, A.L, Turnbull, A.P, Bunkoczi, G, Papagrigoriou, E, Gorrec, F, Umeano, C, Bullock, A, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the bric-a-brac (BTB) domain of human BACH1
To be Published
|
|
2IHD
 
 | | Crystal structure of Human Regulator of G-protein signaling 8, RGS8 | | Descriptor: | CHLORIDE ION, Regulator of G-protein signaling 8 | | Authors: | Turnbull, A.P, Papagrigoriou, E, Ugochukwu, E, Salah, E, Gileadi, C, Burgess, N, Bhatia, C, Gileadi, O, Bray, J, Elkins, J, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2IHE
 
 | | Crystal structure of wild-type single-stranded DNA binding protein from Thermus aquaticus | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Fedorov, R, Witte, G, Urbanke, C, Manstein, D.J, Curth, U. | | Deposit date: | 2006-09-26 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins.
Nucleic Acids Res., 34, 2006
|
|
2IHF
 
 | | Crystal structure of deletion mutant delta 228-252 R190A of the single-stranded DNA binding protein from Thermus aquaticus | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Fedorov, R, Witte, G, Urbanke, C, Manstein, D.J, Curth, U. | | Deposit date: | 2006-09-26 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins.
Nucleic Acids Res., 34, 2006
|
|
2IHJ
 
 | |
2IHK
 
 | |
2IHL
 
 | | LYSOZYME (E.C.3.2.1.17) (JAPANESE QUAIL) | | Descriptor: | JAPANESE QUAIL EGG WHITE LYSOZYME, SODIUM ION | | Authors: | Houdusse, A, Bentley, G.A, Poljak, R.J, Souchon, H, Zhang, Z. | | Deposit date: | 1993-06-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Three-dimensional structure of a heteroclitic antigen-antibody cross-reaction complex.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
2IHM
 
 | | Polymerase mu in ternary complex with gapped 11mer DNA duplex and bound incoming nucleotide | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', 5'-D(*CP*GP*GP*CP*AP*AP*TP*AP*CP*TP*G)-3', ... | | Authors: | Moon, A.F, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2006-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate specificity of DNA Polymerase mu.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2IHN
 
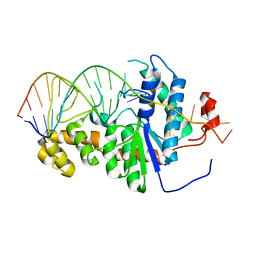 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
