6AOU
 
 | |
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6AOV
 
 | |
3H77
 
 | | Crystal structure of Pseudomonas aeruginosa PqsD in a covalent complex with anthranilate | | Descriptor: | Anthraniloyl-coenzyme A, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
5TK8
 
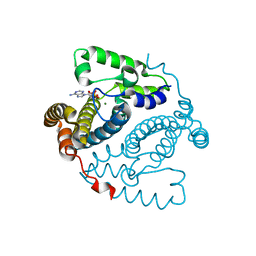 | |
1BPS
 
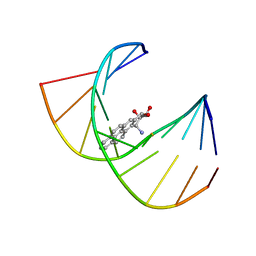 | | MINOR CONFORMER OF A BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA (5'-D(*CP*TP*CP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*(BAP)AP*CP*GP*AP*G)-3') | | Authors: | Schwartz, J.S, Rice, J.S, Luxon, B.A, Sayer, J.M, Xie, G, Yeh, H.J.C, Liu, X, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the minor conformer of a DNA duplex containing a dG mismatch opposite a benzo[a]pyrene diol epoxide/dA adduct: glycosidic rotation from syn to anti at the modified deoxyadenosine.
Biochemistry, 36, 1997
|
|
6AOT
 
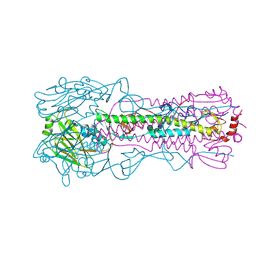 | |
1M3Z
 
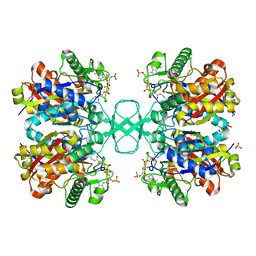 | | Biosynthetic thiolase, C89A mutant, complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
5TK7
 
 | |
1GHE
 
 | | CRYSTAL STRUCTURE OF TABTOXIN RESISTANCE PROTEIN COMPLEXED WITH AN ACYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, ACETYLTRANSFERASE | | Authors: | He, H, Ding, Y, Bartlam, M, Sun, F, Le, Y, Qin, X, Tang, H, Zhang, R, Joachimiak, A, Liu, Y, Zhao, N, Rao, Z. | | Deposit date: | 2000-12-13 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Tabtoxin Resistance Protein Complexed with Acetyl
Coenzyme A Reveals the Mechanism for beta-Lactam Acetylation
J.Mol.Biol., 325, 2003
|
|
5TK6
 
 | |
6BKQ
 
 | |
4DFB
 
 | | Crystal structure of aminoglycoside phosphotransferase aph(2")-id/aph(2")-iva in complex with kanamycin | | Descriptor: | APH(2")-Id, CHLORIDE ION, KANAMYCIN A | | Authors: | Stogios, P.J, Minasov, G, Osipiuk, J, Evdokimova, E, Egorova, E, Di leo, R, Li, H, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A small molecule discrimination map of the antibiotic resistance kinome.
Chem.Biol., 18, 2011
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4HFU
 
 | | Crystal structure of Fab 8M2 in complex with a H2N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 8M2 heavy chain, Fab 8M2 light chain, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5TUS
 
 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4ZE9
 
 | | Se-PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | ABC transporter, substrate binding protein (Agrocinopines A and B), alpha-D-gulopyranose-(1-2)-4-O-phosphono-beta-D-fructofuranose, ... | | Authors: | El Sahili, A, Guimaraes, B.G, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
1UZ9
 
 | | Crystallographic and solution studies of N-lithocholyl insulin: a new generation of prolonged-acting insulins. | | Descriptor: | (2S)-2-AMINO-6-({(4R)-4-[(10R,13S)-10,13-DIMETHYL-3-OXOHEXADECAHYDRO-1H-CYCLOPENTA[A]PHENANTHREN-17-YL]PENTANOYL}AMINO)HEXANOIC ACID, CHLORIDE ION, INSULIN, ... | | Authors: | Whittingham, J.L, Jonassen, I, Havelund, S, Roberts, S.M, Dodson, E.J, Verma, C.S, Wilkinson, A.J, Dodson, G.G. | | Deposit date: | 2004-03-08 | | Release date: | 2005-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and Solution Studies of N-Lithocholyl Insulin: A New Generation of Prolonged-Acting Human Insulins
Biochemistry, 43, 2004
|
|
4ZE8
 
 | | PBP AccA from A. tumefaciens C58 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4ZEC
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocin 84 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4ZEB
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
1N71
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii in complex with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Ghuman, N, Wybenga-Groot, L.E, Berghuis, A.M. | | Deposit date: | 2002-11-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the AAC(6')-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members
Protein Sci., 12, 2003
|
|
3I55
 
 | | Co-crystal structure of Mycalamide A Bound to the Large Ribosomal Subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Steitz, T.A, Moore, P.B. | | Deposit date: | 2009-07-03 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui.
Antimicrob.Agents Chemother., 53, 2009
|
|
4HG4
 
 | | Crystal structure of Fab 2G1 in complex with a H2N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2G1 heavy chain, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-10-06 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4FFS
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Helicobacter pylori with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Haapalainen, A.M, Rinaldo-Matthis, A, Brown, R.L, Norris, G.E, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Picomolar Transition State Analogue Inhibitor of MTAN as a Specific Antibiotic for Helicobacter pylori.
Biochemistry, 51, 2012
|
|
