8HL1
 
 | |
1QZ9
 
 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
1RAR
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, CHLORIDE ION, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
1FFK
 
 | | CRYSTAL STRUCTURE OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Ban, N, Nissen, P, Hansen, J, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.
Science, 289, 2000
|
|
1FCX
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARGAMMA-SELECTIVE RETINOID BMS184394 | | Descriptor: | 6-[HYDROXY-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALEN-2-YL)-METHYL]-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1FCY
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1FCZ
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE PANAGONIST RETINOID BMS181156 | | Descriptor: | 4-[3-OXO-3-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-PROPENYL]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
8HKZ
 
 | |
8HKY
 
 | |
8HKU
 
 | |
8HL2
 
 | |
8HL3
 
 | |
8HKV
 
 | |
8H7E
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) at 2.0 angstrom resolution | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H7C
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) - Pt derivative | | Descriptor: | ACETATE ION, CHLORIDE ION, De novo ferric enterobactin esterase Syn-F4, ... | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H7D
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HL5
 
 | |
8HL4
 
 | |
3RAR
 
 | |
3T0R
 
 | |
1XAP
 
 | | Structure of the ligand binding domain of the Retinoic Acid Receptor beta | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Retinoic acid receptor beta | | Authors: | Germain, P, Kammerer, S, Peluso-Iltis, C, Tortolani, D, Zusi, F.C, Starrett, J, Lapointe, P, Daris, J.P, Marinier, A, De Lera, A.R, Rochel, N, Gronemeyer, H. | | Deposit date: | 2004-08-26 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational design of RAR-selective ligands revealed by RARbeta crystal structure
Embo Rep., 5, 2004
|
|
7RD1
 
 | | The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25 | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Baker, A.T, Boyd, R.J, Sarkar, D, Vermaas, J.V, Williams, D, Singharoy, A. | | Deposit date: | 2021-07-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome.
Sci Adv, 7, 2021
|
|
2OK9
 
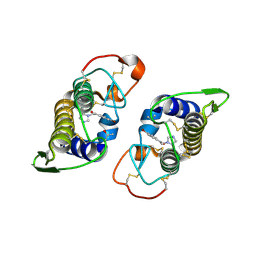 | | PrTX-I-BPB | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog 1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R. | | Deposit date: | 2007-01-16 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a phospholipase A(2) homolog complexed with p-bromophenacyl bromide reveals important structural changes associated with the inhibition of myotoxic activity.
Biochim.Biophys.Acta, 1794, 2009
|
|
2QOG
 
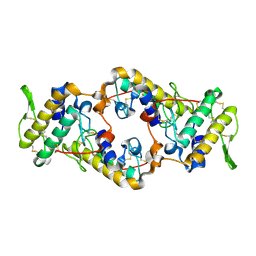 | | Crotoxin B, the basic PLA2 from Crotalus durissus terrificus. | | Descriptor: | CALCIUM ION, Phospholipase A2 CB1, Phospholipase A2 CB2 | | Authors: | Marchi-Salvador, D.P, Correa, L.C, Fontes, M.R.M. | | Deposit date: | 2007-07-20 | | Release date: | 2008-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Insights into the role of oligomeric state on the biological activities of crotoxin: crystal structure of a tetrameric phospholipase A2 formed by two isoforms of crotoxin B from Crotalus durissus terrificus venom.
Proteins, 72, 2008
|
|
7OP2
 
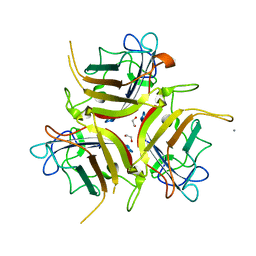 | | Chadox1/ Chimpanzee adenovirus Y25 fiber knob protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Baker, A.T, Parker, A.L, Teijeira Crespo, A, Lipka-Lloyd, M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome.
Sci Adv, 7, 2021
|
|
