7CSY
 
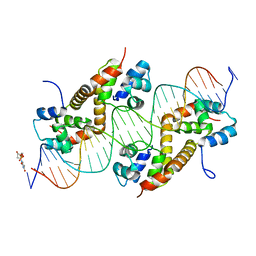 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
6NWZ
 
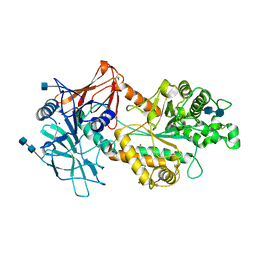 | | Crystal structure of Agd3 a novel carbohydrate deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation.
Nat Commun, 11, 2020
|
|
4XR5
 
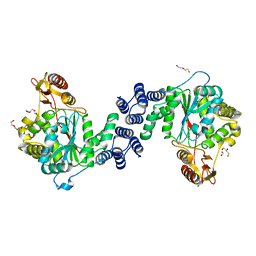 | | X-ray structure of the unliganded thymidine phosphorylase from Salmonella typhimurium at 2.05 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5MJD
 
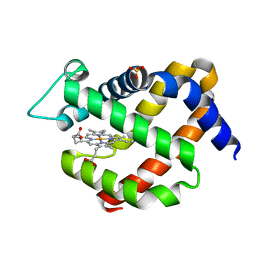 | | metNgb under oxygen at 80 bar | | Descriptor: | Neuroglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5MJC
 
 | | metNeuroglobin under oxygen at 50 bar | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, OXYGEN MOLECULE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
4YGV
 
 | | Reversal Agent for Dabigatran | | Descriptor: | GLYCEROL, aDabi-Fab2a heavy chain, aDabi-Fab2a light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-26 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
7YQA
 
 | | Crystal structure of D-threonine aldolase from Chlamydomonas reinhardtii | | Descriptor: | D-threonine aldolase, MAGNESIUM ION | | Authors: | Hirato, Y, Goto, M, Mizobuchi, T, Muramatsu, H, Tanigawa, M, Nishimura, K. | | Deposit date: | 2022-08-05 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of pyridoxal 5'-phosphate-bound D-threonine aldolase from Chlamydomonas reinhardtii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
4YHM
 
 | | Reversal Agent for Dabigatran | | Descriptor: | N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, aDabi-Fab2b heavy chain, aDabi-Fab2b light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YHI
 
 | | Reversal Agent for Dabigatran | | Descriptor: | N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, aDabi-Fab2a heavy chain, aDabi-Fab2a light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YHO
 
 | | Reversal Agent for Dabigatran | | Descriptor: | N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, aDabi-Fab3 heavy chain, aDabi-Fab3 light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YHK
 
 | | Reversal Agent for Dabigatran | | Descriptor: | GLYCEROL, N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, aDabi-Fab2a heavy chain, ... | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YHL
 
 | | Reversal Agent for Dabigatran | | Descriptor: | aDabi-Fab2b heavy chain, aDabi-Fab2b light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
8FWA
 
 | | Phycocyanin structure from a modular droplet injector for serial femtosecond crystallography | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Botha, S, Doppler, D.L, Sonker, M, Egatz-Gomez, A, Grieco, A, Zaare, S, Jernigan, R, Meza-Aguilar, J.D, Rabbani, M.T, Manna, A, Alvarez, R, Karpos, K, Cruz Villarreal, J, Nelson, G, Ketawala, G.K, Pey, A.L, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Nazari, R, Sierra, R, Hunter, M.S, Batyuk, A, Kupitz, C.J, Sublett, R.E, Lisova, S, Mariani, V, Boutet, S, Fromme, R, Grant, T.D, Fromme, P, Kirian, R.A, Martin-Garcia, J.M, Ros, A. | | Deposit date: | 2023-01-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
4QN1
 
 | | Crystal Structure of a Functionally Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Walker, J.R, Guan, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a Function Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH
To be Published
|
|
6Q5N
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24Nle | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CC-Hex*-L24Nle, GLYCEROL | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6PWY
 
 | | Structure of C. elegans ZK177.8, SAMHD1 ortholog | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lim, C.S, Maehigashi, T, Wade, L.R, Bowen, N, Knecht, K, Xiong, Y, Kim, B. | | Deposit date: | 2019-07-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | ZK177.8 of Caenorhabditis elegans:
Aicardi-Goutieres Syndrome SAMHD1 Ortholog
To Be Published
|
|
5OHJ
 
 | | Human phosphodiesterase 4B catalytic domain in complex with a pyrrolidinyl inhibitor. | | Descriptor: | MAGNESIUM ION, ZINC ION, [(1~{S})-2-[3,5-bis(chloranyl)-1-oxidanyl-pyridin-1-ium-4-yl]-1-[4-[bis(fluoranyl)methoxy]-3-(cyclopropylmethoxy)phenyl]ethyl] (2~{S})-1-[3-(dimethylcarbamoyl)phenyl]sulfonylpyrrolidine-2-carboxylate, ... | | Authors: | Rizzi, A, Carzaniga, L, Armani, E. | | Deposit date: | 2017-07-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Thiazolidinyl and Pyrrolidinyl Derivatives as Inhaled PDE4 Inhibitors for Respiratory Diseases.
J. Med. Chem., 60, 2017
|
|
8J30
 
 | | Crystal structure of ApNGT with Q469A and M218A mutations in complex with UDP-GLC | | Descriptor: | UDP-glucose:protein N-beta-glucosyltransferase, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Feng, Y, Hao, Z, Guo, Q, Zheng, J, Da, L, Peng, W. | | Deposit date: | 2023-04-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Investigation of the Catalytic Mechanism of a Soluble N-glycosyltransferase Allows Synthesis of N-glycans at Noncanonical Sequons.
Jacs Au, 3, 2023
|
|
6Q5R
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL-KgEb | | Descriptor: | CC-Hex*-LL-KgEb, GLYCEROL | | Authors: | Beesley, J.L, Rhys, G.G, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5H
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24D | | Descriptor: | AMMONIUM ION, CC-Hex*-L24D, SULFATE ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5K
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24K | | Descriptor: | AMMONIUM ION, CC-Hex*-L24K, GLYCEROL, ... | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5Q
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex-KgEb | | Descriptor: | CC-Hex-KgEb, L(+)-TARTARIC ACID | | Authors: | Zaccai, N.R, Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5J
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24E | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24E | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5M
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24Dab | | Descriptor: | CC-Hex*-L24Dab, SODIUM ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
7Q7K
 
 | | JAK2 in complex with 4-(2-amino-8-methoxyquinazolin-6-yl)phenol | | Descriptor: | 4-(2-azanyl-8-methoxy-quinazolin-6-yl)phenol, Tyrosine-protein kinase JAK2 | | Authors: | Rowland, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Investigation of Janus Kinase (JAK) Inhibitors for Lung Delivery and the Importance of Aldehyde Oxidase Metabolism.
J.Med.Chem., 65, 2022
|
|
