6XUP
 
 | |
6XV0
 
 | | lauric acid functionalized hexamolybdoaluminate bound to human serum albumin | | Descriptor: | MYRISTIC ACID, Serum albumin, lauric acid functionalized hexamolybdoaluminate | | Authors: | Bijelic, A, Dobrov, A, Roller, A, Rompel, A. | | Deposit date: | 2020-01-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a Fatty Acid-Functionalized Anderson-Type Polyoxometalate to Human Serum Albumin.
Inorg.Chem., 59, 2020
|
|
7BXZ
 
 | |
8G3M
 
 | |
6XWV
 
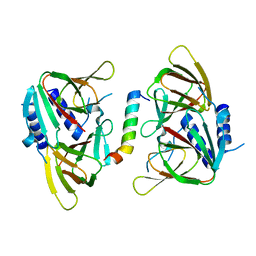 | | Crystal structure of drosophila melanogaster CENP-C bound to CAL1 | | Descriptor: | Calmodulin, Ryanodine Receptor 2 | | Authors: | Jeyaprakash, A.A, Medina-Pritchard, B, Lazou, V, Zou, J, Byron, O, Abad, M.A, Rappsilber, J, Heun, P. | | Deposit date: | 2020-01-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for centromere maintenance by Drosophila CENP-A chaperone CAL1.
Embo J., 39, 2020
|
|
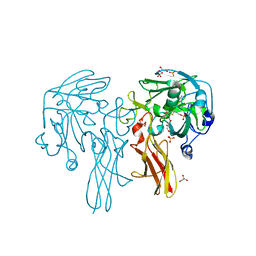
6Y0K
 
 | |
8GBX
 
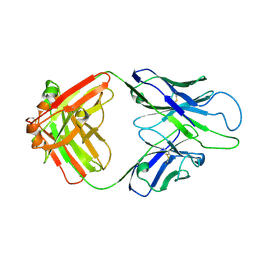 | |
6MZJ
 
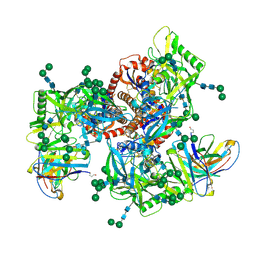 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6OPQ
 
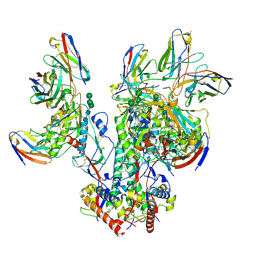 | | CD4- and 17-bound HIV-1 Env B41 SOSIP frozen with LMNG | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6O8S
 
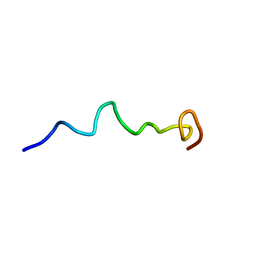 | | Syn-safencin 56 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
8GBY
 
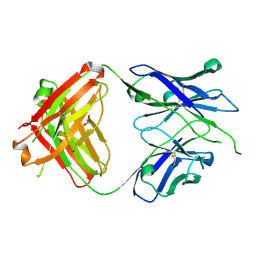 | | Crystal structure of PC39-50E, an anti-HIV broadly neutralizing antibody | | Descriptor: | GLYCEROL, PC39-50E Fab heavy chain, PC39-50E Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GBW
 
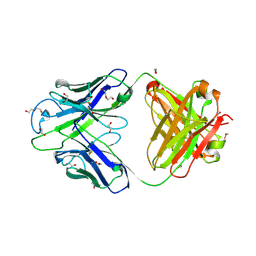 | | Crystal structure of PC39-23D, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PC39-23D Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GC0
 
 | | Crystal structure of PC39-50L, an anti-HIV broadly neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PC39-50L Fab heavy chain, PC39-50L Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GBV
 
 | | Crystal structure of PC39-17A, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PC39-17A Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
6N8H
 
 | |
7PDT
 
 | | Crystal structure of a mutated form of RXRalpha ligand binding domain in complex with BMS649 and a coactivator fragment | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W, Guee, L. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
6MV3
 
 | | NMR structure of the cNTnC-cTnI chimera bound to calcium desensitizer W7 | | Descriptor: | CALCIUM ION, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE, Troponin C, ... | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2018-10-24 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Changes Induced by the Binding of the Calcium Desensitizer W7 to Cardiac Troponin.
Biochemistry, 57, 2018
|
|
6QDA
 
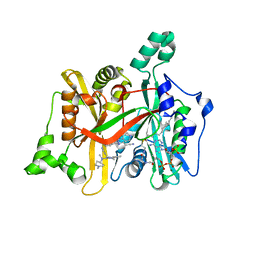 | | Leishmania major N-myristoyltransferase in complex with quinazoline inhibitor IMP-0000811 | | Descriptor: | 3-[methyl-[2-[methyl-(1-methylpiperidin-4-yl)amino]quinazolin-4-yl]amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDG
 
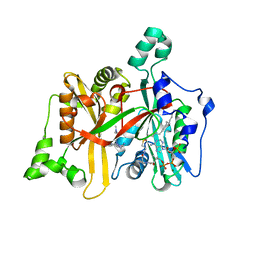 | | Leishmania major N-myristoyltransferase in complex with quinazoline inhibitor IMP-0000169 | | Descriptor: | 3-[[6-bromanyl-2-[3-(dimethylamino)propyl-methyl-amino]quinazolin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
8U1Q
 
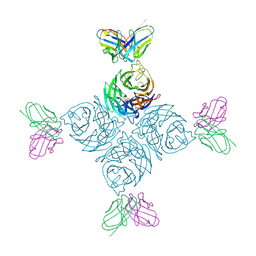 | |
8U1S
 
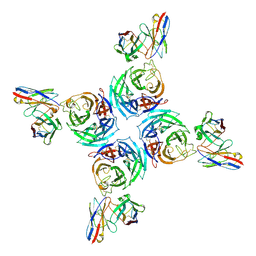 | |
6NB4
 
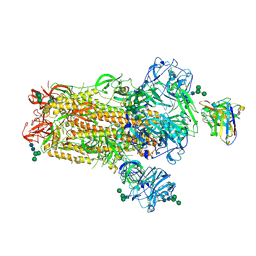 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
7PZB
 
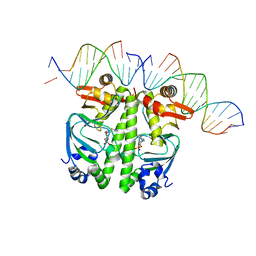 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
7PZW
 
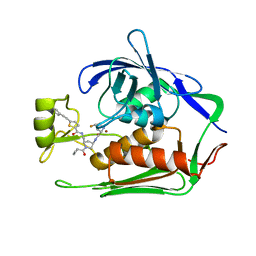 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (4S)-N-((3R,5R)-1-(cyclopropanecarbonyl)-5-((5-(phenylethynyl)thiophene-2-carboxamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
8EF3
 
 | |
