5J9L
 
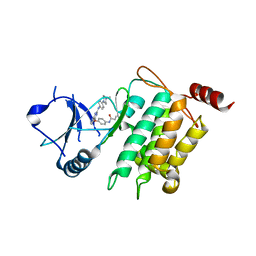 | | Crystal structure of CPT1691 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, N-(4-((2-((4-(4-methylpiperazin-1-yl)phenyl)amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy)phenyl)acrylamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7515 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
6HSK
 
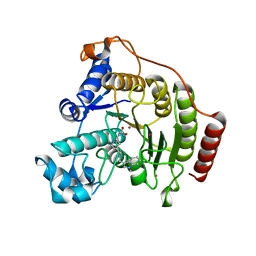 | | Crystal structure of a human HDAC8 L6 loop mutant complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Ramos-Morales, E, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
5JFO
 
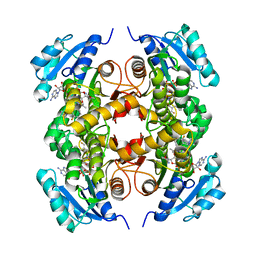 | | Structure of the M.tuberculosis enoyl-reductase InhA in complex with GSK625 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-{1-[(2-chloro-6-fluorophenyl)methyl]-1H-pyrazol-3-yl}-5-[(1S)-1-(3-methyl-1H-pyrazol-1-yl)ethyl]-1,3,4-thiadiazol-2-amine, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gulten, G, Sacchettini, J.C. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Antitubercular drugs for an old target: GSK693 as a promising InhA direct inhibitor.
Ebiomedicine, 8, 2016
|
|
5KZ5
 
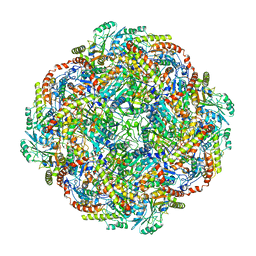 | | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Complex Formed by the Iron Donor, the Sulfur Donor, and the Scaffold | | Descriptor: | Cysteine desulfurase, mitochondrial, Frataxin, ... | | Authors: | Gakh, O, Ranatunga, W, Smith, D.Y, Ahlgren, E.C, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery.
J.Biol.Chem., 291, 2016
|
|
7DFR
 
 | |
4RSR
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with trivalent phenyl arsencial derivative-Roxarsone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-arsanyl-2-nitrophenol, Arsenic methyltransferase, ... | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6RT9
 
 | |
7YJD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
5WKO
 
 | |
9IVV
 
 | | Crystal structure of human secretory glutaminyl cyclase in complex with the inhibitor 3-((2-(1H-imidazol-5-yl)ethyl)carbamoyl)-4-amino-1,2,5-oxadiazole 2-oxide (compound 13) | | Descriptor: | 4-azanyl-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-2-oxidanidyl-1,2,5-oxadiazol-2-ium-3-carboxamide, DIMETHYL SULFOXIDE, DIMETHYLFORMAMIDE, ... | | Authors: | Li, G.-B, Yu, J.-L, Zhou, C, Ning, X.-L, Mou, J, Wu, J.-W, Meng, F.-B. | | Deposit date: | 2024-07-24 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.961 Å) | | Cite: | Knowledge-guided diffusion model for 3D ligand-pharmacophore mapping.
Nat Commun, 16, 2025
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
6BAX
 
 | |
5U9I
 
 | | Crystal structure of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6APU
 
 | | Crystal structure of Trypanosoma brucei hypoxanthine-guanine phosphoribosyltranferase in complex with (2-{[2-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](3-aminopropyl)amino}ethyl)phosphonic acid | | Descriptor: | (2-{[2-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)ethyl](3-aminopropyl)amino}ethyl)phosphonic acid, DI(HYDROXYETHYL)ETHER, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Teran, D, Guddat, L. | | Deposit date: | 2017-08-18 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Evaluation of the Trypanosoma brucei 6-oxopurine salvage pathway as a potential target for drug discovery.
PLoS Negl Trop Dis, 12, 2018
|
|
1LDY
 
 | |
6IB7
 
 | | Structure of wild type SuhB | | Descriptor: | GLYCEROL, Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
4TYJ
 
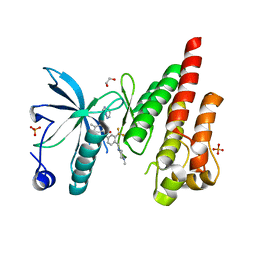 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Fibroblast growth factor receptor 4, ... | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
6C9N
 
 | |
9AT5
 
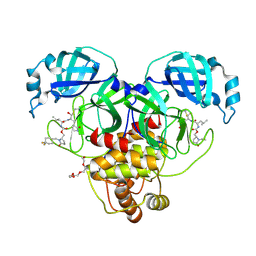 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATF
 
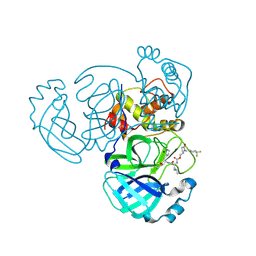 | | Crystal structure of MERS 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
6EPR
 
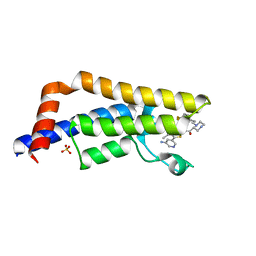 | | The ATAD2 bromodomain in complex with compound UZH-DS15 | | Descriptor: | (~{N}~{Z},2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-3~{H}-1,3-thiazol-2-ylidene]-1-[2-[4,4-bis(fluoranyl)cyclohexyl]ethyl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Hitting a moving target: simulation and crystallography study of ATAD2 bromodomain blockers
To Be Published
|
|
5YQ5
 
 | | Crystal structure of human osteomodulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Osteomodulin, PHOSPHATE ION | | Authors: | Caaveiro, J.M.M, Tashima, T, Tsumoto, K. | | Deposit date: | 2017-11-05 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular basis for governing the morphology of type-I collagen fibrils by Osteomodulin.
Commun Biol, 1, 2018
|
|
6Q5F
 
 | | OXA-48_P68A-CAZ. Evolutionary trade-offs of OXA-48 mediated ceftazidime-avibactam resistance | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Frohlich, C, Sorum, V, Thomassen, A.M, Johnsen, P.J, Leiros, H.-K.S, Samuelsen, O. | | Deposit date: | 2018-12-08 | | Release date: | 2019-03-20 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | OXA-48-Mediated Ceftazidime-Avibactam Resistance Is Associated with Evolutionary Trade-Offs.
Msphere, 4, 2019
|
|
5TCK
 
 | | Second Bromodomain from Leishmania donovani LdBPK.091320 complexed with Bromosporine | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromosporine, ... | | Authors: | Lin, L.-H, Hou, C.F.D, Loppnau, P, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-09 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Second Bromodomain from Leishmania donovani LdBPK.091320 complexed with Bromosporine
To Be Published
|
|
8BXL
 
 | | Patulin Synthase from Penicillium expansum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tjallinks, G, Boverio, A, Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2022-12-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure elucidation and characterization of patulin synthase, insights into the formation of a fungal mycotoxin.
Febs J., 290, 2023
|
|
