2MPF
 
 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MNG
 
 | | Apo Structure of human HCN4 CNBD solved by NMR | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | Deposit date: | 2014-04-03 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
2MHF
 
 | |
2KXL
 
 | |
2K0G
 
 | |
2HKX
 
 | | Structure of CooA mutant (N127L/S128L) from Carboxydothermus hydrogenoformans | | Descriptor: | CARBON MONOXIDE, Carbon monoxide oxidation system transcription regulator CooA-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lanz, N.D, Borjigin, M, Li, H, Kerby, R.L, Poulos, T.L, Roberts, G.P. | | Deposit date: | 2006-07-05 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2H6C
 
 | |
2H6B
 
 | |
2GZW
 
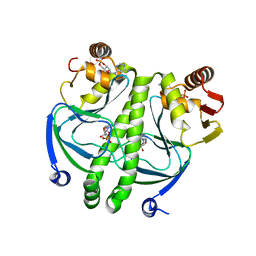 | | Crystal structure of the E.coli CRP-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Kumarevel, T.S, Tanaka, T, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of activated CRP protein from E coli
To be Published
|
|
2GAU
 
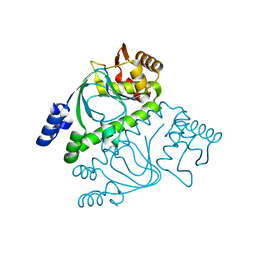 | | Crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis (APC80792), Structural genomics, MCSG | | Descriptor: | transcriptional regulator, Crp/Fnr family | | Authors: | Rotella, F.J, Zhang, R.G, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis
To be Published
|
|
2FMY
 
 | |
2D93
 
 | |
2CGP
 
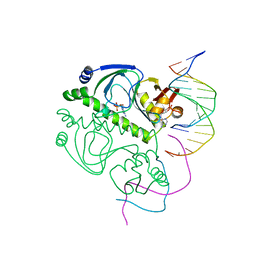 | | CATABOLITE GENE ACTIVATOR PROTEIN/DNA COMPLEX, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*AP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*TP*TP*AP*AP*T)-3'), ... | | Authors: | Passner, J.M, Steitz, T.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2BYV
 
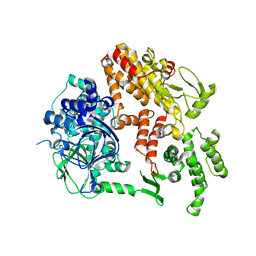 | |
1ZYB
 
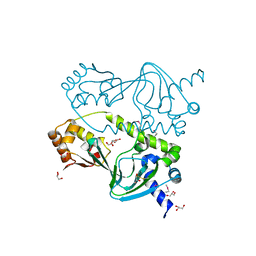 | |
1ZRF
 
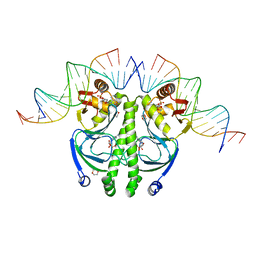 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6C;17G]ICAP38 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRE
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6G;17C]ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*GP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*CP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRD
 
 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6A;17T]ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*AP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*TP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1ZRC
 
 | | 4 Crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-ICAP38 DNA | | Descriptor: | 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
1VP6
 
 | | M.loti ion channel cylic nucleotide binding domain | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, BROMIDE ION, Cyclic-nucleotide binding domain of mesorhizobium loti CNG potassium channel | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-10-14 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel.
Cell(Cambridge,Mass.), 119, 2004
|
|
1U12
 
 | | M. loti cyclic nucleotide binding domain mutant | | Descriptor: | IODIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Clayton, G.M, Silverman, W.R, Heginbotham, L, Morais-Cabral, J.H. | | Deposit date: | 2004-07-14 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Ligand Activation in a Cyclic Nucleotide Regulated Potassium Channel
Cell(Cambridge,Mass.), 119, 2004
|
|
1RUO
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1RL3
 
 | | Crystal structure of cAMP-free R1a subunit of PKA | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Brown, S, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2003-11-24 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RIalpha subunit of PKA: a cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B.
Structure, 12, 2004
|
|
1RGS
 
 | | REGULATORY SUBUNIT OF CAMP DEPENDENT PROTEIN KINASE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP DEPENDENT PROTEIN KINASE | | Authors: | Su, Y, Dostmann, W.R.G, Herberg, F.W, Durick, K, Xuong, N.-H, Ten Eyck, L, Taylor, S.S, Varughese, K.I. | | Deposit date: | 1995-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains.
Science, 269, 1995
|
|
