4D9Y
 
 | | The crystal structure of Chelerythrine bound to DNA d(CGTACG) | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, CALCIUM ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Chelerythrine bound to DNA d(CGTACG)
To be Published
|
|
9FF6
 
 | | Human transthyretin (TTR) in complex with (E)-4-((((2-methoxybenzyl)oxy)imino)methyl)benzoic acid (Lic157) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(~{Z})-(2-methoxyphenyl)methoxyiminomethyl]benzoic acid, GLYCEROL, ... | | Authors: | Ciccone, L, Shepard, W, Sirigu, S, Camodeca, C, Mazzoccchi, F, Fruchart, C, Nencetti, S, Orlandini, E. | | Deposit date: | 2024-05-22 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human transthyretin (TTR) in complex with (E)-4-((((2-methoxybenzyl)oxy)imino)methyl)benzoic acid (Lic157)
To Be Published
|
|
5BJU
 
 | | X-ray structure of the PglF dehydratase from Campylobacter jejuni in complex with UDP and NAD(H) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
6MTZ
 
 | |
1TNK
 
 | |
8CUR
 
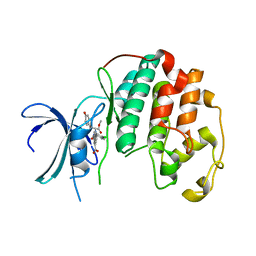 | | Crystal structure of Cdk2 in complex with Cyclin A inhibitor 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid | | Descriptor: | 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid, Cyclin-dependent kinase 2 | | Authors: | Tripathi, S.M, Tambo, C.S, Kiss, G, Rubin, S.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biolayer Interferometry Assay for Cyclin-Dependent Kinase-Cyclin Association Reveals Diverse Effects of Cdk2 Inhibitors on Cyclin Binding Kinetics.
Acs Chem.Biol., 18, 2023
|
|
9FMO
 
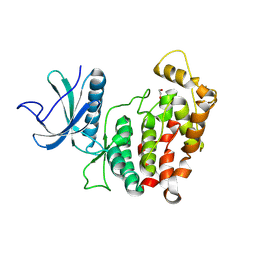 | | Crystal structure of C. merolae LAMMER-like dual specificity kinase (CmLIK) kinase domain | | Descriptor: | 1,2-ETHANEDIOL, LAMMER-like dual specificity kinase, ZINC ION | | Authors: | Dimos-Roehl, B, Haltenhof, T, Kotte, A, Heyd, F, Loll, B. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Crystal structure of C. merolae LAMMER-like dual specificity kinase CmLIK kinase domain
to be published
|
|
8WFW
 
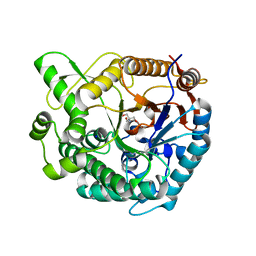 | |
6FR0
 
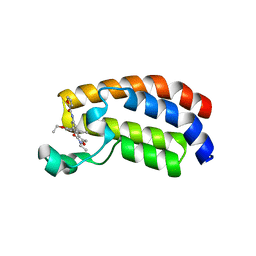 | | Crystal structure of CREBBP bromodomain complexd with PB08 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(2-ethyl-5-methyl-3-oxidanylidene-1,2-oxazol-4-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FRF
 
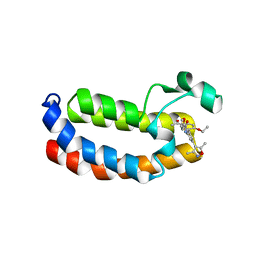 | | Crystal structure of CREBBP bromodomain complexd with PA10 | | Descriptor: | CREB-binding protein, ~{N}-[3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
8WFV
 
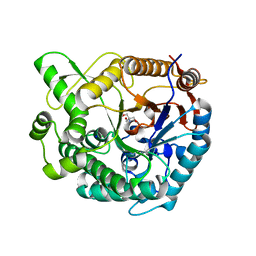 | |
6STT
 
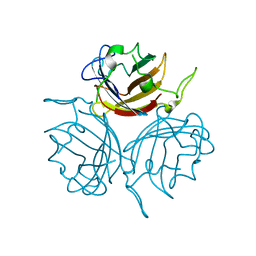 | | Adenovirus 29 Fiber Knob protein in complex with Sialic acid | | Descriptor: | Fiber, N-acetyl-alpha-neuraminic acid | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2019-09-11 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus
npj Viruses, 1, 2023
|
|
6FQZ
 
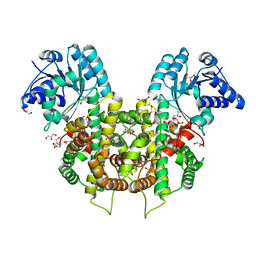 | | Plasmodium falciparum 6-phosphogluconate dehydrogenase in its apo form, in complex with its cofactor NADP+ and in complex with its substrate 6-phosphogluconate | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconate dehydrogenase, decarboxylating, ... | | Authors: | Fritz-Wolf, K, Haeussler, K, Reichmann, M, Rahlfs, S, Becker, K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of Plasmodium falciparum 6-Phosphogluconate Dehydrogenase as an Antimalarial Drug Target.
J. Mol. Biol., 430, 2018
|
|
8WA5
 
 | |
8QT2
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-6 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
7T8G
 
 | |
5F5Y
 
 | |
5LXI
 
 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
5J9W
 
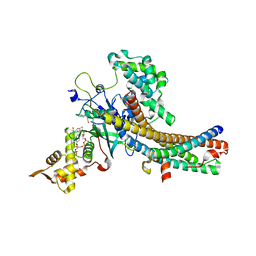 | | Crystal structure of the NuA4 core complex | | Descriptor: | ACETYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
4B7H
 
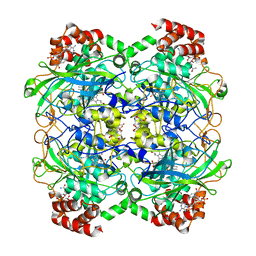 | | Structure of a highdose liganded bacterial catalase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATALASE, CHLORIDE ION, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
9LFV
 
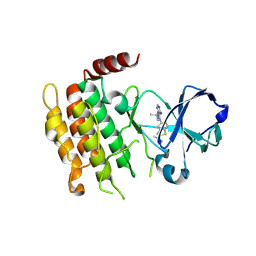 | | Crystal structure of mouse RIP3 kinase domain complexed with LK01004 | | Descriptor: | 3-[2-[(2~{R})-butan-2-yl]-1,3-benzothiazol-5-yl]-1-~{tert}-butyl-pyrazolo[3,4-d]pyrimidin-4-amine, 3-[2-[(2~{S})-butan-2-yl]-1,3-benzothiazol-5-yl]-1-~{tert}-butyl-pyrazolo[3,4-d]pyrimidin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2025-01-09 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure-based design of potent and selective inhibitors targeting RIPK3 for eliminating on-target toxicity in vitro.
Nat Commun, 16, 2025
|
|
5ZRN
 
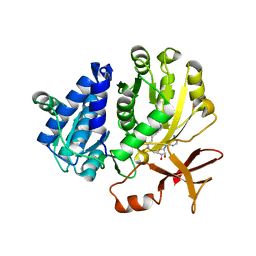 | |
9D1Q
 
 | | Lucilia cuprina alpha esterase 7 directed evolution round 6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Carboxylic ester hydrolase, ... | | Authors: | Frkic, R.L, Fraser, N, Mabbitt, P.D, Jackson, C.J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lucilia cuprina alpha esterase 7 directed evolution round 6
To Be Published
|
|
8HP0
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
9D1R
 
 | | Lucilia cuprina alpha esterase 7 directed evolution round 7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Carboxylic ester hydrolase, ... | | Authors: | Frkic, R.L, Fraser, N, Mabbitt, P.D, Jackson, C.J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Lucilia cuprina alpha esterase 7 directed evolution round 7
To Be Published
|
|
