2ARK
 
 | | Structure of a flavodoxin from Aquifex aeolicus | | Descriptor: | Flavodoxin, GLYCEROL, PHOSPHATE ION | | Authors: | Cuff, M.E, Quartey, P, Zhou, M, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a flavodoxin from Aquifex aeolicus
To be Published
|
|
1V1M
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-20 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain Alpha-Keto Acid Decarboxylase/Dehydrogenase.
J.Biol.Chem., 279, 2004
|
|
1UZH
 
 | | A CHIMERIC CHLAMYDOMONAS, SYNECHOCOCCUS RUBISCO ENZYME | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Karkehabadi, S, Spreitzer, R.J, Andersson, I. | | Deposit date: | 2004-03-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chimeric Small Subunits Influence Catalysis without Causing Global Conformational Changes in the Crystal Structure of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Biochemistry, 44, 2005
|
|
2B1W
 
 | |
1VLG
 
 | |
2C9D
 
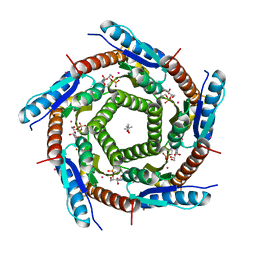 | | Lumazine Synthase from Mycobacterium tuberculosis Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL)HEXANE 1-PHOSPHATE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL ) HEXANE 1-PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
2CE9
 
 | |
2CAJ
 
 | | NikR from Helicobacter pylori in closed trans-conformation and nickel bound to 4 intermediary sites | | Descriptor: | CHLORIDE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
1W21
 
 | | Structure of Neuraminidase from English duck subtype N6 complexed with 30 mM sialic acid (NANA, Neu5Ac), crystal soaked for 43 hours at 291 K. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2004-06-25 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Crystal Structure of Type a Influenza Virus Neuraminidase of the N6 Subtype Reveals the Existence of Two Separate Neu5Ac Binding Sites
To be Published
|
|
2ATY
 
 | | Complement receptor chimaeric conjugate CR2-Ig | | Descriptor: | Complement receptor chimeric conjugate CR2-Ig | | Authors: | Gilbert, H.E, Aslam, M, Guthridge, J.M, Holers, V.M, Perkins, S.J. | | Deposit date: | 2005-08-26 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Extended Flexible Linker Structures in the Complement Chimaeric Conjugate CR2-Ig by Scattering, Analytical Ultracentrifugation and Constrained Modelling: Implications for Function and Therapy.
J.Mol.Biol., 356, 2006
|
|
2ARO
 
 | | Crystal Structure Of The Native Histone Octamer To 2.1 Angstrom Resolution, Crystalised In The Presence Of S-Nitrosoglutathione | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Sodngam, S, Nicholson, J.M, Lambert, S.J, Reynolds, C.D, Baldwin, J.P. | | Deposit date: | 2005-08-20 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The oxidised histone octamer does not form a H3 disulphide bond.
Biochim.Biophys.Acta, 1764, 2006
|
|
2AXT
 
 | | Crystal Structure of Photosystem II from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Loll, B, Kern, J, Saenger, W, Zouni, A, Biesiadka, J. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-27 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II
NATURE, 438, 2005
|
|
1XWN
 
 | | solution structure of cyclophilin like 1(PPIL1) and insights into its interaction with SKIP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase like 1 | | Authors: | Xu, C, Xu, Y, Tang, Y, Wu, J, Shi, Y, Huang, Q, Zhang, Q. | | Deposit date: | 2004-11-01 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human peptidyl prolyl isomerase like protein 1 and insights into its interaction with SKIP
J.Biol.Chem., 281, 2006
|
|
1XNQ
 
 | |
1XSQ
 
 | | Crystal structure of ureidoglycolate hydrolase from E.coli. Northeast Structural Genomics Consortium target ET81. | | Descriptor: | Ureidoglycolate hydrolase | | Authors: | Kuzin, A.P, Vorobiev, S.M, Abashidze, M, Acton, T.B, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-19 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase from E.coli. Northeast Structural Genomics Consortium target ET81.
To be Published
|
|
2CML
 
 | | Structure of Neuraminidase from English Duck Subtype N6 Complexed with 30 MM ZANAMIVIR, Crystal Soaked for 3 Hours at 291 K. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Lukacik, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2006-05-10 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Type a Influenza Virus Neuraminidase of the N6 Subtype Reveals the Existence of Two Separate Neu5Ac Binding Sites
To be Published
|
|
2C97
 
 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 4-(6- chloro-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)butyl phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(6-CHLORO-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL) BUTYL PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
1XWE
 
 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
1XNR
 
 | |
1XXX
 
 | | Crystal structure of Dihydrodipicolinate Synthase (DapA, Rv2753c) from Mycobacterium tuberculosis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, Dihydrodipicolinate synthase, ... | | Authors: | Kefala, G, Panjikar, S, Janowski, R, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-09 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure and kinetic study of dihydrodipicolinate synthase from Mycobacterium tuberculosis.
Biochem.J., 411, 2008
|
|
1XFV
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Q, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XMQ
 
 | | Crystal Structure of t6A37-ASLLysUUU AAA-mRNA Bound to the Decoding Center | | Descriptor: | 16s ribosomal RNA, 30S Ribosomal Protein S10, 30S Ribosomal Protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XFW
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3'5' cyclic AMP (cAMP) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XHV
 
 | | HincII bound to cleaved cognate DNA GTCGAC and Mn2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*C)-3', 5'-D(P*GP*AP*CP*CP*GP*G)-3', MANGANESE (II) ION, ... | | Authors: | Etzkorn, C, Horton, N.C. | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Insights from the Structures of HincII Bound to Cognate DNA Cleaved from Addition of Mg(2+) and Mn(2+)
J.Mol.Biol., 343, 2004
|
|
1XK4
 
 | | Crystal structure of human calprotectin(S100A8/S100A9) | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Korndoerfer, I.P, Brueckner, F, Skerra, A. | | Deposit date: | 2004-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the human (S100A8/S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of interacting alpha-helices can determine specific association of two EF-hand proteins
J.Mol.Biol., 370, 2007
|
|
