2FLV
 
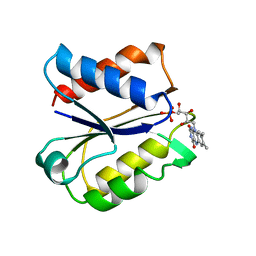 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57T REDUCED (150K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-19 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
2FLW
 
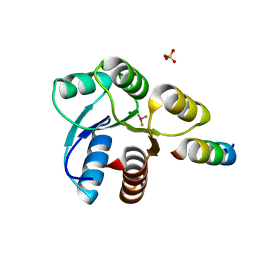 | | Crystal structure of Mg2+ and BeF3- ound CheY in complex with CheZ 200-214 solved from a F432 crystal grown in Hepes (pH 7.5) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, Chemotaxis protein cheY, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation.
J.Mol.Biol., 359, 2006
|
|
2FLY
 
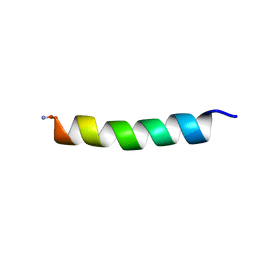 | | Proadrenomedullin N-Terminal 20 Peptide | | Descriptor: | Proadrenomedullin N-20 terminal peptide | | Authors: | Lucyk, S, Taha, H, Yamamoto, H, Miskolzie, M, Kotovych, G. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR conformational analysis of proadrenomedullin N-terminal 20 peptide, a proangiogenic factor involved in tumor growth
Biopolymers, 81, 2006
|
|
2FLZ
 
 | |
2FM0
 
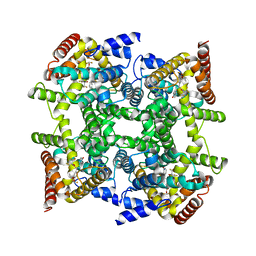 | | Crystal structure of PDE4D in complex with L-869298 | | Descriptor: | (S)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHY L)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2FM1
 
 | |
2FM2
 
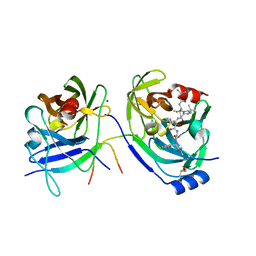 | | HCV NS3-4A protease domain complexed with a ketoamide inhibitor, SCH446211 | | Descriptor: | BETA-MERCAPTOETHANOL, NS3 protease/helicase, NS4a protein, ... | | Authors: | Yi, M, Tong, X, Skelton, A, Chase, R, Chen, T, Prongay, A, Bogen, S.L, Saksena, A.K, Njoroge, F.G, Veselenak, R.L, Pyles, R.B, Bourne, N, Malcolm, B.A, Lemon, S.M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations
J.Biol.Chem., 281, 2006
|
|
2FM4
 
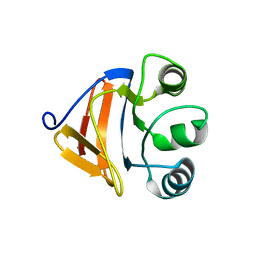 | |
2FM5
 
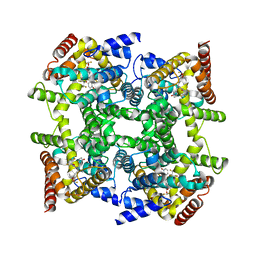 | | Crystal structure of PDE4D2 in complex with inhibitor L-869299 | | Descriptor: | (R)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHYL)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-07 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2FM6
 
 | | Zinc-beta-lactamase L1 from stenotrophomonas maltophilia (native form) | | Descriptor: | GLYCEROL, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-01-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
2FM7
 
 | |
2FM8
 
 | |
2FM9
 
 | |
2FMA
 
 | |
2FMB
 
 | | EIAV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, EQUINE INFECTIOUS ANEMIA VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
2FMC
 
 | |
2FMD
 
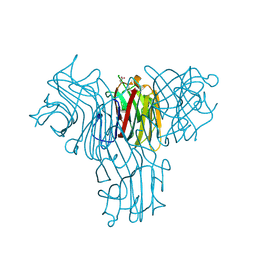 | | Structural basis of carbohydrate recognition by Bowringia milbraedii seed agglutinin | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Buts, L, Garcia-Pino, A, Wyns, L, Loris, R. | | Deposit date: | 2006-01-09 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of carbohydrate recognition by a Man(alpha1-2)Man-specific lectin from Bowringia milbraedii.
Glycobiology, 16, 2006
|
|
2FME
 
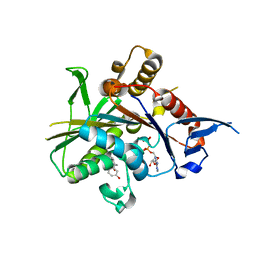 | | Crystal structure of the mitotic kinesin eg5 (ksp) in complex with mg-adp and (r)-4-(3-hydroxyphenyl)-n,n,7,8-tetramethyl-3,4-dihydroisoquinoline-2(1h)-carboxamide | | Descriptor: | (4R)-4-(3-HYDROXYPHENYL)-N,N,7,8-TETRAMETHYL-3,4-DIHYDROISOQUINOLINE-2(1H)-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Sheriff, S. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of human mitotic kinesin Eg5: Characterization of the 4-phenyl-tetrahydroisoquinoline lead series
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FMF
 
 | |
2FMG
 
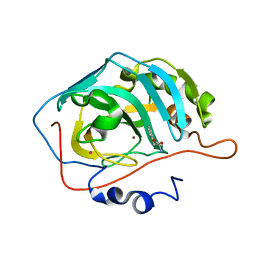 | | Carbonic anhydrase activators. Activation of isoforms I, II, IV, VA, VII and XIV with L- and D- phenylalanine and crystallographic analysis of their adducts with isozyme II: sterospecific recognition within the active site of an enzyme and its consequences for the drug design, structure with L-phenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, PHENYLALANINE, ... | | Authors: | Temperini, C, Scozzafava, A, Vullo, D, Supuran, C.T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbonic Anhydrase Activators. Activation of Isoforms I, II, IV, VA, VII, and XIV with l- and d-Phenylalanine and Crystallographic Analysis of Their Adducts with Isozyme II: Stereospecific Recognition within the Active Site of an Enzyme and Its Consequences for the Drug Design.
J.Med.Chem., 49, 2006
|
|
2FMH
 
 | | Crystal structure of Mg2+ and BeF3- bound CheY in complex with CheZ 200-214 solved from a F432 crystal grown in Tris (pH 8.4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation.
J.Mol.Biol., 359, 2006
|
|
2FMI
 
 | | Crystal structure of CheY in complex with CheZ 200-214 solved from a F432 crystal grown in Tris (pH 8.4) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-terminal 15-mer from Chemotaxis protein cheZ, Chemotaxis protein cheY, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation.
J.Mol.Biol., 359, 2006
|
|
2FMJ
 
 | |
2FMK
 
 | | Crystal structure of Mg2+ and BeF3- bound CheY in complex with CheZ 200-214 solved from a P2(1)2(1)2 crystal grown in MES (pH 6.0) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, C-terminal 15-mer from Chemotaxis protein cheZ, Chemotaxis protein cheY, ... | | Authors: | Guhaniyogi, J, Robinson, V.L, Stock, A.M. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal Structures of Beryllium Fluoride-free and Beryllium Fluoride-bound CheY in Complex with the Conserved C-terminal Peptide of CheZ Reveal Dual Binding Modes Specific to CheY Conformation
J.Mol.Biol., 359, 2006
|
|
2FML
 
 | |
