4UEQ
 
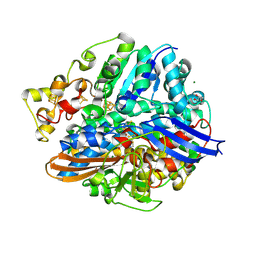 | | Structure of the V74C large subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CALCIUM ION, CARBONATE ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
4UE6
 
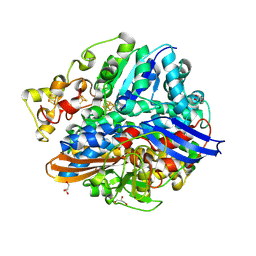 | | Structure of methylene blue-treated anaerobically purified D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | [Nife]-Hydrogenases Revisited: Nickel-Carboxamido Bond Formation in a Variant with Accrued O2-Tolerance and a Tentative Re-Interpretation of Ni-Si States.
Metallomics, 7, 2015
|
|
4UEW
 
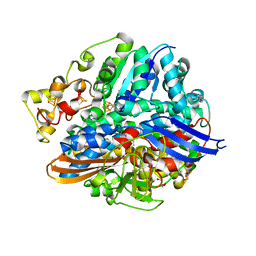 | | Structure of H2-treated anaerobically purified D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
4CJ9
 
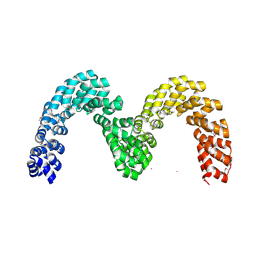 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in its apo form | | Descriptor: | BURRH, SELENIUM ATOM | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6F48
 
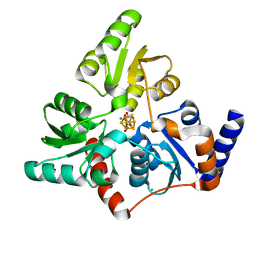 | | Structure of quinolinate synthase with reaction intermediates X and Y | | Descriptor: | 2-imino,3-carboxy,5-hydroxy,6-oxo hexanoic acid, 5-hydroxy,-4,5-dihydroquinolinate, CHLORIDE ION, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Trapping of Reaction Intermediates in Quinolinic Acid Synthesis by NadA.
ACS Chem. Biol., 13, 2018
|
|
4DCX
 
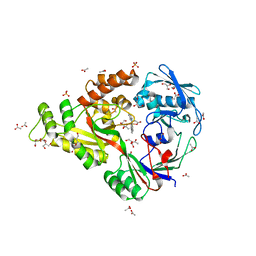 | | X-ray structure of NikA in complex with Fe(1R,2R)-N,N'-Bis(2-pyridylmethyl)-N,N'-dicarboxymethyl-1,2-cyclohexanediamine | | Descriptor: | ACETATE ION, GLYCEROL, Nickel-binding periplasmic protein, ... | | Authors: | Cherrier, M.V, Girgenti, E, Amara, P, Iannello, M, Marchi-Delapierre, C, Fontecilla-Camps, J.C, Menage, S, Cavazza, C. | | Deposit date: | 2012-01-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the periplasmic nickel-binding protein NikA provides insights for artificial metalloenzyme design.
J.Biol.Inorg.Chem., 17, 2012
|
|
6F4L
 
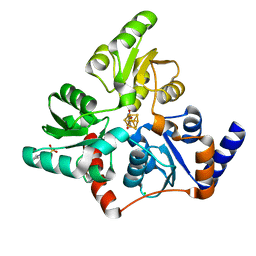 | |
9C59
 
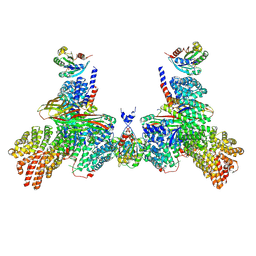 | |
8VY3
 
 | | Human DNA polymerase alpha/primase - AavLEA1 (1:40 molar ratio) | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Abe, K.M, Li, G, Grant, T, Lim, C.J. | | Deposit date: | 2024-02-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Small LEA proteins mitigate air-water interface damage to fragile cryo-EM samples during plunge freezing.
Nat Commun, 15, 2024
|
|
8UCE
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
3QAL
 
 | | Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
1TW9
 
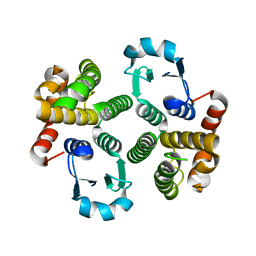 | | Glutathione Transferase-2, apo form, from the nematode Heligmosomoides polygyrus | | Descriptor: | glutathione S-transferase 2 | | Authors: | Schuller, D.J, Liu, Q, Kriksunov, I.A, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of a new class of glutathione transferase from the model human hookworm nematode Heligmosomoides polygyrus.
Proteins, 61, 2005
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
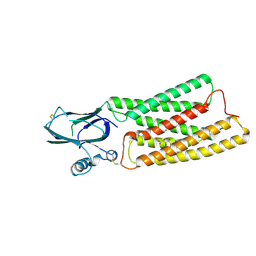 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
1TTF
 
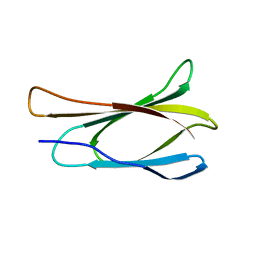 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
7RWK
 
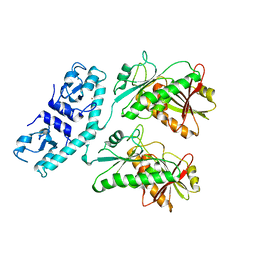 | | Structure of Cap5 from Asticcacaulis sp. | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein, ZINC ION | | Authors: | Huang, R.H, Fatma, S, Chakravarti, A. | | Deposit date: | 2021-08-20 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular mechanisms of the CdnG-Cap5 antiphage defense system employing 3',2'-cGAMP as the second messenger.
Nat Commun, 12, 2021
|
|
7RWM
 
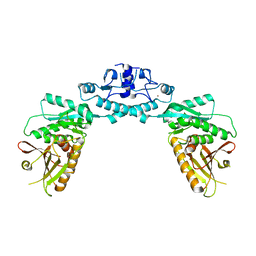 | | Structure of Cap5 from Lactococcus lactis | | Descriptor: | SAVED domain-containing protein, ZINC ION | | Authors: | Huang, R.H, Fatma, S, Chakravarti, A. | | Deposit date: | 2021-08-20 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular mechanisms of the CdnG-Cap5 antiphage defense system employing 3',2'-cGAMP as the second messenger.
Nat Commun, 12, 2021
|
|
1TTG
 
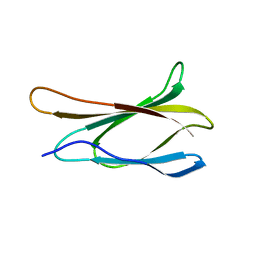 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
3S4P
 
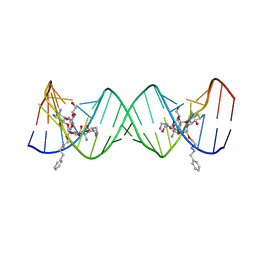 | | Crystal structure of the bacterial ribosomal decoding site complexed with an amphiphilic paromomycin O2''-ether analogue | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-2-O-{2-[(2-phenylethyl)amino]ethyl}-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Szychowski, J, Kondo, J, Zahr, O, Auclair, K, Westhof, E, Hanessian, S, Keillor, J.W. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Inhibition of aminoglycoside-deactivating enzymes APH(3')-IIIa and AAC(6')-Ii by amphiphilic paromomycin O2''-ether analogues
Chemmedchem, 6, 2011
|
|
7OHF
 
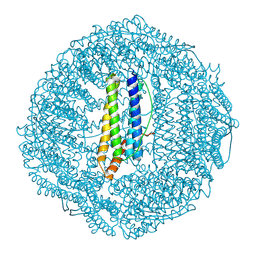 | | Cryo-EM structure of pyrococcus furiosus apoferritin in nanofluidic channels | | Descriptor: | Ferritin | | Authors: | Huber, S.T, Sarajlic, E, Huijink, R, Evers, W.H, Jakobi, A.J. | | Deposit date: | 2021-05-10 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Nanofluidic chips for cryo-EM structure determination from picoliter sample volumes.
Elife, 11, 2022
|
|
4UUC
 
 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
4V35
 
 | | The Structure of A-PGS from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CALCIUM ION, ... | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V34
 
 | | The Structure of A-PGS from Pseudomonas aeruginosa (SeMet derivative) | | Descriptor: | ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CHLORIDE ION, SULFATE ION | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KET
 
 | | Structure of the aldo-keto reductase from Coptotermes gestroi | | Descriptor: | Aldo-keto reductase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liberato, M.L, Campos, B.M, Tramontina, R, Squina, F.M. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Coptotermes gestroi aldo-keto reductase: a multipurpose enzyme for biorefinery applications.
Biotechnol Biofuels, 10, 2017
|
|
