8S51
 
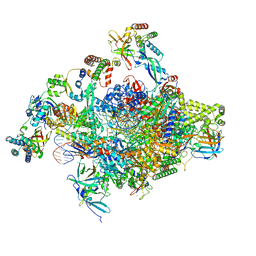 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S52
 
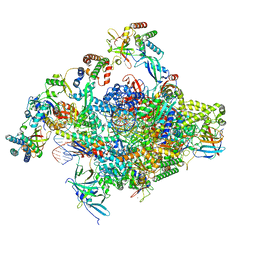 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
7P2D
 
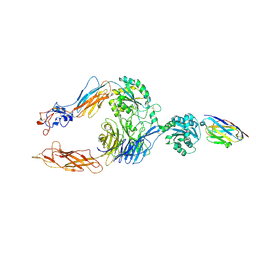 | | Structure of alphaMbeta2/Cd11bCD18 headpiece in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2021-07-05 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
J.Biol.Chem., 298, 2022
|
|
8S54
 
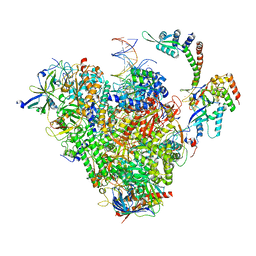 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
7PFM
 
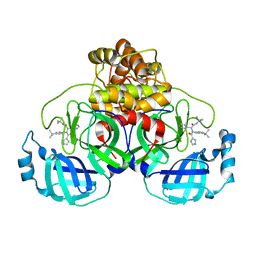 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
7PHE
 
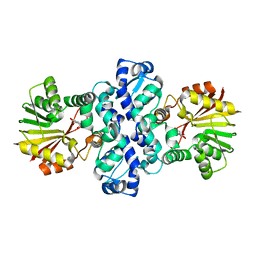 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7P25
 
 | |
7P3U
 
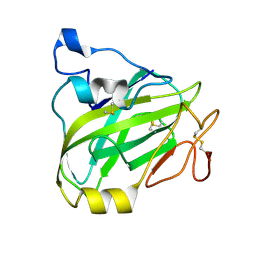 | | Chitin-active fungal AA11 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, putative | | Authors: | Rohr, A.K, Stoepamo, F.G, Eijsink, V.G.H. | | Deposit date: | 2021-07-08 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of a lytic polysaccharide monooxygenase from Aspergillus fumigatus shows functional variation among family AA11 fungal LPMOs.
J.Biol.Chem., 297, 2021
|
|
8RG0
 
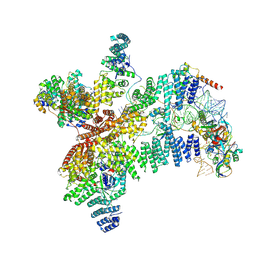 | | Structure of human eIF3 core from closed 48S translation initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7PBF
 
 | |
7PXQ
 
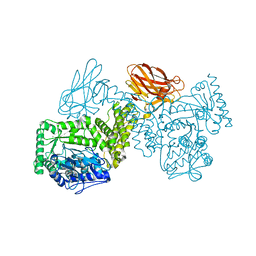 | | GH115 alpha-1,2-glucuronidase D303A | | Descriptor: | CALCIUM ION, xylan alpha-1,2-glucuronidase | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5HHT
 
 | |
7MDH
 
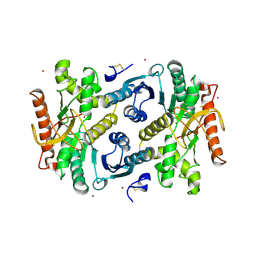 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
7Q96
 
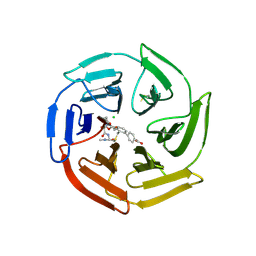 | | Keap1 compound complex | | Descriptor: | 4-[(5S,8R)-5-(dimethylcarbamoyl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-trien-16-yl]benzoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7Q6S
 
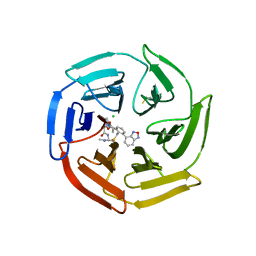 | | Keap1 compound complex | | Descriptor: | (5S,8R)-16-(2,1,3-benzoxadiazol-4-yl)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-7,11-bis(oxidanylidene)-10-oxa-3-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
5HLO
 
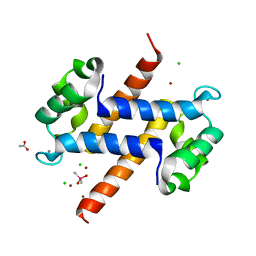 | | Crystal structure of calcium and zinc-bound human S100A8 in space group C2221 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lin, H, Andersen, G.R, Yatime, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human S100A8 in complex with zinc and calcium.
Bmc Struct.Biol., 16, 2016
|
|
7QB4
 
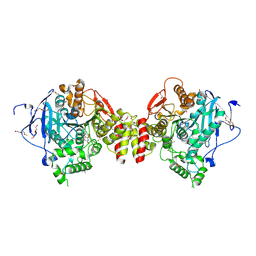 | | Mus Musculus Acetylcholinesterase in complex with 7-[(1-benzylpiperidin-3-yl)methoxy]-3,4-dimethyl-2H-chromen-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ekstrom, F.J, Forsgren, N. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.500011 Å) | | Cite: | Dual Reversible Coumarin Inhibitors Mutually Bound to Monoamine Oxidase B and Acetylcholinesterase Crystal Structures.
Acs Med.Chem.Lett., 13, 2022
|
|
7QAK
 
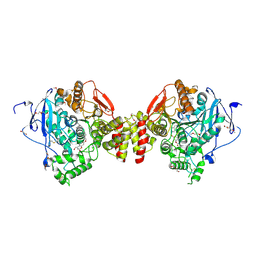 | | Mus Musculus Acetylcholinesterase in complex with 7-[(4-{[benzyl(methyl)amino]methyl}benzyl)oxy]-4-(hydroxymethyl)-2H-chromen-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Ekstrom, F.J, Forsgren, N. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.60000324 Å) | | Cite: | Dual Reversible Coumarin Inhibitors Mutually Bound to Monoamine Oxidase B and Acetylcholinesterase Crystal Structures.
Acs Med.Chem.Lett., 13, 2022
|
|
7Q1Y
 
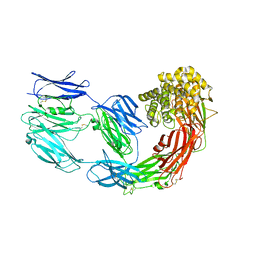 | | X-ray structure of human A2ML1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin-like protein 1, ... | | Authors: | Andersen, G.R, Zarantonello, A, Enghild, J.J, Nielsen, N.S. | | Deposit date: | 2021-10-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Cryo-EM structures of human A2ML1 elucidate the protease-inhibitory mechanism of the A2M family.
Nat Commun, 13, 2022
|
|
6V5D
 
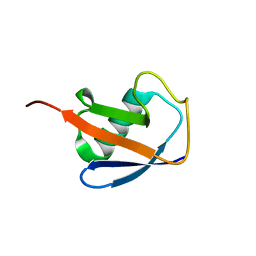 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
7PZ1
 
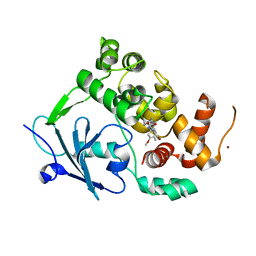 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH8535 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(3-methoxy-4-methyl-phenyl)piperidine-1-carboxamide, GLYCEROL, ... | | Authors: | Scaletti, E.R, Helleday, T, Stenmark, P. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
7Q3K
 
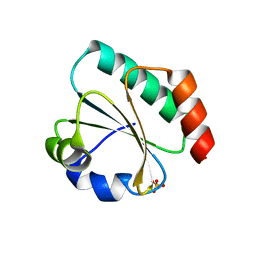 | | Computationally designed thioredoxin subjected to stability optimizing mutations. | | Descriptor: | SULFATE ION, eMM9 | | Authors: | Norrild, R.K, Johansson, K.E, O'Shea, C, Lindorff-Larsen, K, Winther, J.R, Morth, J.P. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Increasing protein stability by inferring substitution effects from high-throughput experiments.
Cell Rep Methods, 2, 2022
|
|
7QFY
 
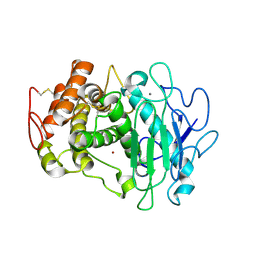 | | Fusarium oxysporum M36 protease without the propeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular metalloproteinase, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fusarium oxysporum M36 protease without the propeptide
To Be Published
|
|
7QLS
 
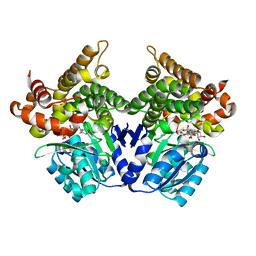 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND DIMETHOXYPHENYL ACETAMIDE | | Descriptor: | 2-(3,4-dimethoxyphenyl)ethanamide, ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
7QLQ
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NAD, AND DIMETHOXYPHENYL ACETAMIDE | | Descriptor: | 2-(3,4-dimethoxyphenyl)ethanamide, ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Sridhar, S, Kiema, T.R, Widersten, M, Wierenga, R.K. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
