9I8V
 
 | | Cryo-EM structure of the Danio rerio tRNA ligase complex | | Descriptor: | ATP-dependent RNA helicase, Ashwin, Family with sequence similarity 98, ... | | Authors: | Chamera, S, Zajko, W, Czarnocki-Cieciura, M, Jaciuk, M, Koziej, L, Nowak, J, Wycisk, K, Sroka, M, Chramiec-Glabik, A, Smietanski, M, Golebiowski, F, Warminski, M, Jemielity, J, Glatt, S, Nowotny, M. | | Deposit date: | 2025-02-06 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural and biochemical characterization of the 3'-5' tRNA splicing ligases.
J.Biol.Chem., 301, 2025
|
|
4WVU
 
 | | CRYSTAL STRUCTURE OF XIAP-BIR2 DOMAIN COMPLEXED WITH LIGAND BOUND | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
4WVT
 
 | | Crystal structure of XIAP-BIR2 domain complexed with ligand bound | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
6E06
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in citrate precipitant) | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-07-06 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6E05
 
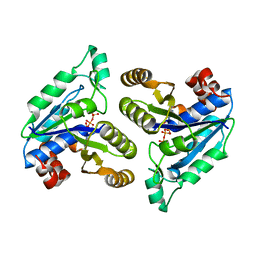 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with cytidine triphosphate solved by precipitant-ligand exchange (crystals grown in sulfate precipitant) | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-07-06 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7TPJ
 
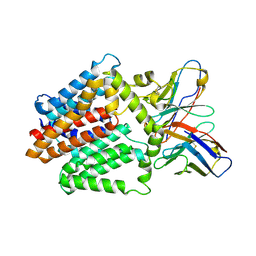 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
3N7Y
 
 | |
4UYL
 
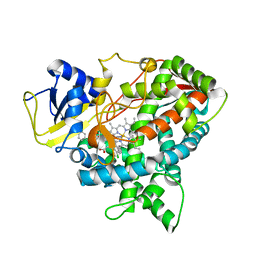 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from a pathogenic filamentous fungus Aspergillus fumigatus in complex with VNI | | Descriptor: | 14-ALPHA STEROL DEMETHYLASE, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2014-09-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-Functional Characterization of Cytochrome P450 Sterol Alpha-Demethylase (Cyp51B) from Aspergillus Fumigatus and Molecular Basis for the Development of Antifungal Drugs
J.Biol.Chem., 290, 2015
|
|
7QBK
 
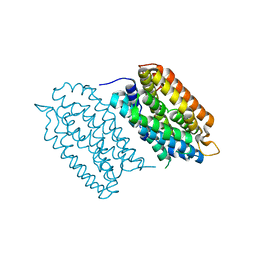 | | Crystal structure of a second homolog of R2-like ligand-binding oxidase in Sulfolobus acidocaldarius (SaR2loxII) | | Descriptor: | FE (III) ION, MANGANESE (III) ION, R2-like ligand-binding oxidase (homolog II) from Sulfolobus acidocaldarius | | Authors: | Lebrette, H, Diamanti, R, Srinivas, V, Hogbom, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Febs Lett., 596, 2022
|
|
7QBP
 
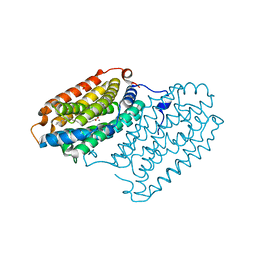 | | Crystal structure of R2-like ligand-binding oxidase from Saccharopolyspora Erythraea | | Descriptor: | FE (III) ION, MANGANESE (III) ION, PALMITIC ACID, ... | | Authors: | Srinivas, V, Diamanti, R, Lebrette, H, Hogbom, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Febs Lett., 596, 2022
|
|
7L34
 
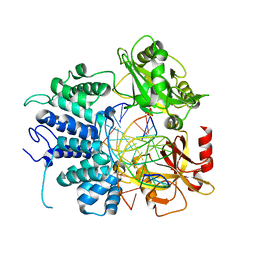 | | Human DNA Ligase 1 - R641L nicked DNA complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Tumbale, P.P, Williams, R.S, Schellenberg, M.S. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | LIG1 syndrome mutations remodel a cooperative network of ligand binding interactions to compromise ligation efficiency.
Nucleic Acids Res., 49, 2021
|
|
4UOU
 
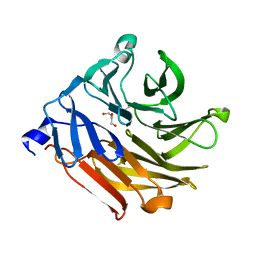 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) - apo-form | | Descriptor: | DI(HYDROXYETHYL)ETHER, FUCOSE-SPECIFIC LECTIN FLEA | | Authors: | Houser, J, Komarek, J, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4USA
 
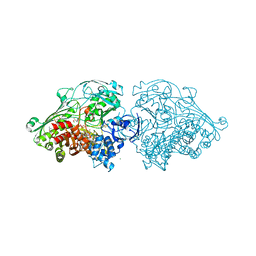 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with trans-cinnamaldehyde | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
7L35
 
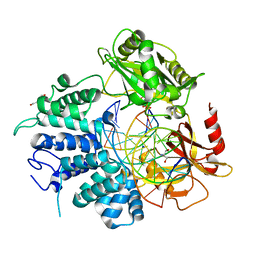 | | Human DNA Ligase 1 - R771W nicked DNA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tumbale, P.P, Williams, R.S, Schellenberg, M.S. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LIG1 syndrome mutations remodel a cooperative network of ligand binding interactions to compromise ligation efficiency.
Nucleic Acids Res., 49, 2021
|
|
4WD3
 
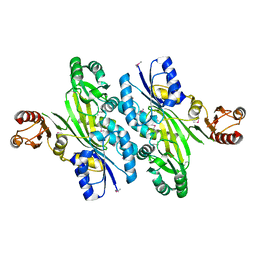 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4UUG
 
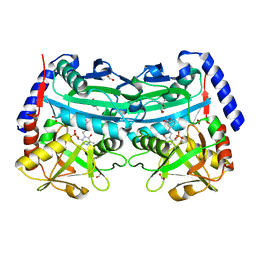 | | The (R)-selective amine transaminase from Aspergillus fumigatus with inhibitor bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Thomsen, M, Hinrichs, W. | | Deposit date: | 2014-07-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Dual Substrate Recognition of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus
FEBS J., 282, 2015
|
|
4V20
 
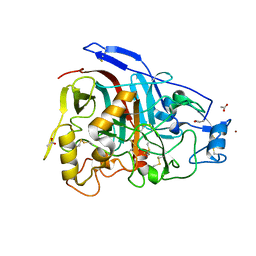 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus, disaccharide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CELLOBIOHYDROLASE, ... | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-05 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7OD1
 
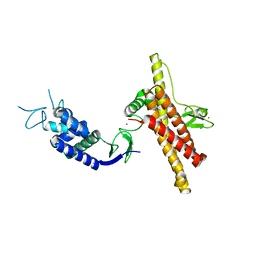 | |
7TZI
 
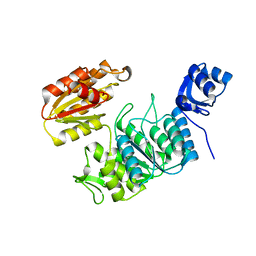 | | Structure of a pseudomurein peptide ligase type E from Methanothermobacter thermautotrophicus | | Descriptor: | Mur ligase family protein | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
8UCH
 
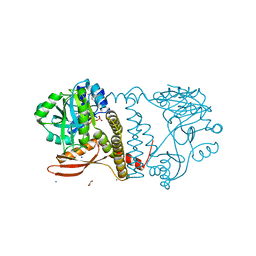 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCE
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCG
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCF
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
4LH7
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-aminothieno[3,2-c]pyridine-2,7-dicarboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Boriack-Sjodin, P.A, Prince, D.B. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
