1SMN
 
 | |
1GBA
 
 | |
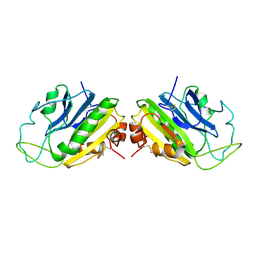
1LFA
 
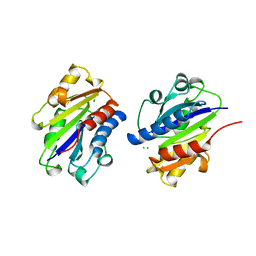 | | CD11A I-DOMAIN WITH BOUND MN++ | | Descriptor: | CD11A, CHLORIDE ION, MANGANESE (II) ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
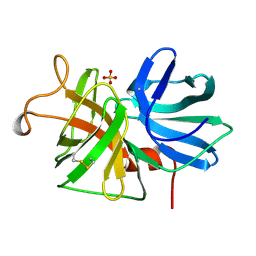
1LMW
 
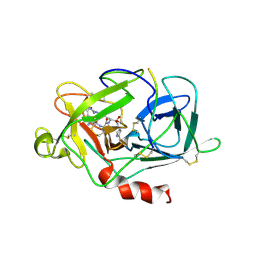 | | LMW U-PA Structure complexed with EGRCMK (GLU-GLY-ARG Chloromethyl Ketone) | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Spraggon, G.S, Phillips, C, Nowak, U.K, Ponting, C.P, Saunders, D, Dobson, C.M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1995-07-26 | | Release date: | 1996-01-29 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the catalytic domain of human urokinase-type plasminogen activator.
Structure, 3, 1995
|
|
1MHC
 
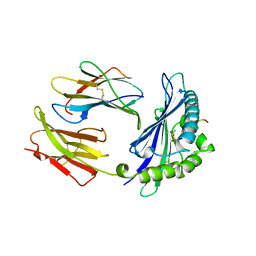 | | MODEL OF MHC CLASS I H2-M3 WITH NONAPEPTIDE FROM RAT ND1 REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I ANTIGEN H2-M3, NONAPEPTIDE FROM RAT NADH DEHYDROGENASE | | Authors: | Wang, C.-R, Fischer Lindahl, K, Deisenhofer, J. | | Deposit date: | 1995-08-23 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3
Cell(Cambridge,Mass.), 82, 1995
|
|
226D
 
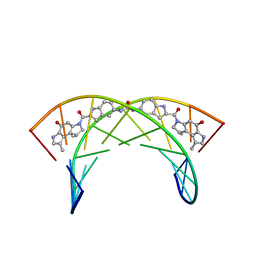 | |
1SYN
 
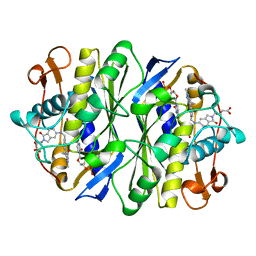 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH BW1843U89 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Stroud, R.M. | | Deposit date: | 1995-09-19 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex of the anti-cancer therapeutic, BW1843U89, with thymidylate synthase at 2.0 A resolution: implications for a new mode of inhibition.
Structure, 4, 1996
|
|
1VAS
 
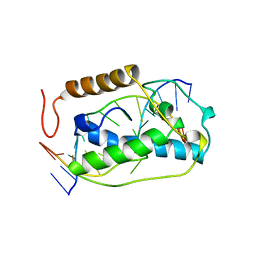 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | Authors: | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
213D
 
 | |
245D
 
 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1996-01-12 | | Release date: | 1996-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1LBG
 
 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
243D
 
 | | STRUCTURE OF THE DNA OCTANUCLEOTIDE D(ACGTACGT)2 | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3') | | Authors: | Wilcock, D.J, Adams, A, Cardin, C.J, Wakelin, L.P.G. | | Deposit date: | 1996-01-10 | | Release date: | 1996-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DNA octanucleotide d(ACGTACGT)2.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1SOL
 
 | | A PIP2 AND F-ACTIN-BINDING SITE OF GELSOLIN, RESIDUE 150-169 (NMR, AVERAGED STRUCTURE) | | Descriptor: | GELSOLIN (150-169) | | Authors: | Xian, W, Vegners, R, Janmey, P.A, Braunlin, W.H. | | Deposit date: | 1995-09-29 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spectroscopic studies of a phosphoinositide-binding peptide from gelsolin: behavior in solutions of mixed solvent and anionic micelles.
Biophys.J., 69, 1995
|
|
1MST
 
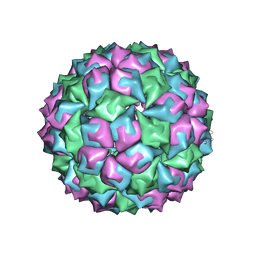 | |
1DCH
 
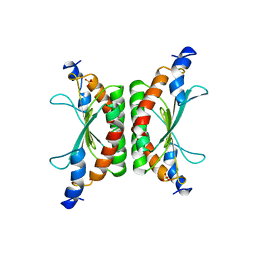 | | CRYSTAL STRUCTURE OF DCOH, A BIFUNCTIONAL, PROTEIN-BINDING TRANSCRIPTION COACTIVATOR | | Descriptor: | DCOH (DIMERIZATION COFACTOR OF HNF-1), SULFATE ION | | Authors: | Endrizzi, J.A, Cronk, J.D, Wang, W, Crabtree, G.R, Alber, T. | | Deposit date: | 1995-01-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator.
Science, 268, 1995
|
|
1DKT
 
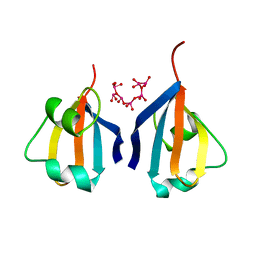 | | CKSHS1: HUMAN CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1 COMPLEX WITH METAVANADATE | | Descriptor: | CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1, META VANADATE | | Authors: | Bourne, Y, Arvai, A.S, Tainer, J.A. | | Deposit date: | 1995-11-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain.
J.Mol.Biol., 249, 1995
|
|
1SPE
 
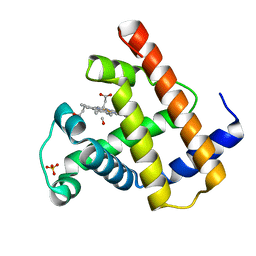 | | SPERM WHALE NATIVE CO MYOGLOBIN AT PH 4.0, TEMP 4C | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1SE2
 
 | |
1DKS
 
 | | CKSHS1: HUMAN CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1 IN COMPLEX WITH PHOSPHATE | | Descriptor: | CYCLIN DEPENDENT KINASE SUBUNIT, TYPE 1, PHOSPHATE ION | | Authors: | Bourne, Y, Arvai, A.S, Tainer, J.A. | | Deposit date: | 1995-11-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain.
J.Mol.Biol., 249, 1995
|
|
1VTP
 
 | |
1NEH
 
 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
1AGG
 
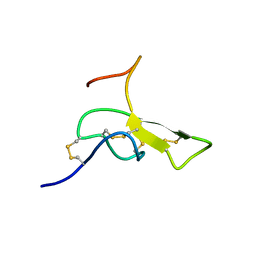 | | THE SOLUTION STRUCTURE OF OMEGA-AGA-IVB, A P-TYPE CALCIUM CHANNEL ANTAGONIST FROM THE VENOM OF AGELENOPSIS APERTA | | Descriptor: | OMEGA-AGATOXIN-IVB | | Authors: | Reily, M.D, Thanabal, V, Adams, M.E. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of omega-Aga-IVB, a P-type calcium channel antagonist from venom of the funnel web spider, Agelenopsis aperta.
J.Biomol.NMR, 5, 1995
|
|
1MLA
 
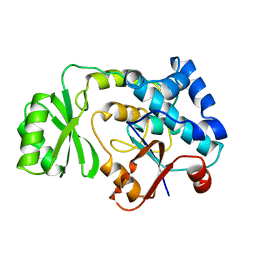 | | THE ESCHERICHIA COLI MALONYL-COA:ACYL CARRIER PROTEIN TRANSACYLASE AT 1.5-ANGSTROMS RESOLUTION. CRYSTAL STRUCTURE OF A FATTY ACID SYNTHASE COMPONENT | | Descriptor: | MALONYL-COENZYME A ACYL CARRIER PROTEIN TRANSACYLASE | | Authors: | Serre, L, Verbree, E.C, Dauter, Z, Stuitje, A.R, Derewenda, Z.S. | | Deposit date: | 1995-01-25 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli malonyl-CoA:acyl carrier protein transacylase at 1.5-A resolution. Crystal structure of a fatty acid synthase component.
J.Biol.Chem., 270, 1995
|
|
2DGC
 
 | |
1QWF
 
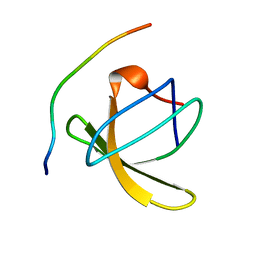 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
